Details of the Target
General Information of Target
| Target ID | LDTP04822 | |||||
|---|---|---|---|---|---|---|
| Target Name | Histone H2B type 1-D (H2BC5) | |||||
| Gene Name | H2BC5 | |||||
| Gene ID | 3017 | |||||
| Synonyms |
H2BFB; HIRIP2; HIST1H2BD; Histone H2B type 1-D; H2B-clustered histone 5; HIRA-interacting protein 2; Histone H2B.1 B; Histone H2B.b; H2B/b |
|||||
| 3D Structure | ||||||
| Sequence |
MPEPTKSAPAPKKGSKKAVTKAQKKDGKKRKRSRKESYSVYVYKVLKQVHPDTGISSKAM
GIMNSFVNDIFERIAGEASRLAHYNKRSTITSREIQTAVRLLLPGELAKHAVSEGTKAVT KYTSSK |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Histone H2B family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Core component of nucleosome. Nucleosomes wrap and compact DNA into chromatin, limiting DNA accessibility to the cellular machineries which require DNA as a template. Histones thereby play a central role in transcription regulation, DNA repair, DNA replication and chromosomal stability. DNA accessibility is regulated via a complex set of post-translational modifications of histones, also called histone code, and nucleosome remodeling.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TH211 Probe Info |
 |
Y122(20.00); Y84(16.42); Y43(10.74); Y41(6.07) | LDD0257 | [1] | |
|
TH214 Probe Info |
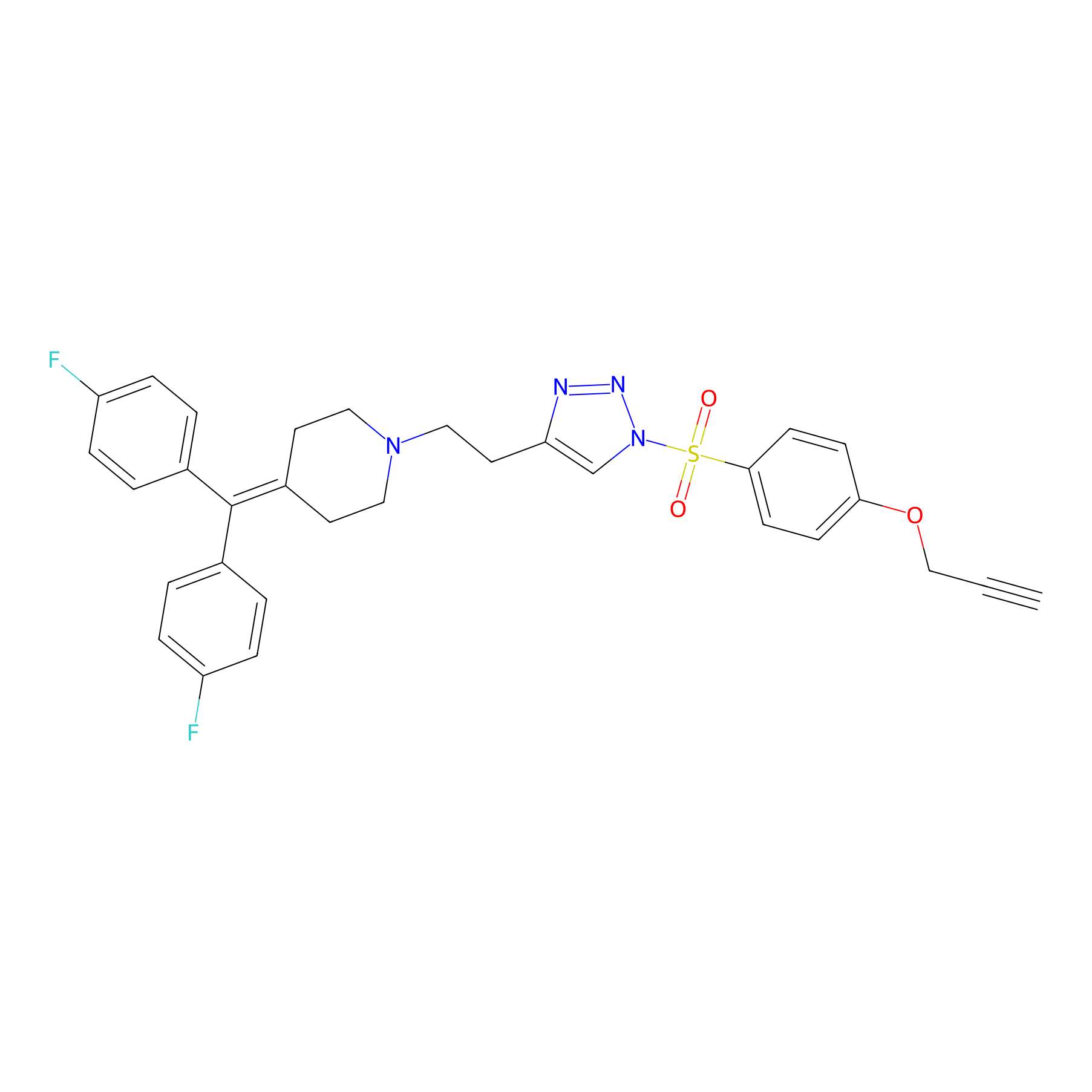 |
Y84(5.49) | LDD0258 | [1] | |
|
TH216 Probe Info |
 |
Y38(20.00); Y43(20.00); Y84(20.00); Y122(16.93) | LDD0259 | [1] | |
|
STPyne Probe Info |
 |
K6(8.07) | LDD0277 | [2] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [2] | |
|
HHS-482 Probe Info |
 |
Y41(0.82); Y43(1.32); Y84(1.51) | LDD0285 | [3] | |
|
HHS-475 Probe Info |
 |
Y84(1.13) | LDD0264 | [4] | |
|
HHS-465 Probe Info |
 |
Y84(2.95) | LDD2237 | [5] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [6] | |
|
SF Probe Info |
 |
Y122(0.00); K6(0.00); K47(0.00); K109(0.00) | LDD0028 | [7] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0116 | HHS-0101 | DM93 | Y84(1.13) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y84(1.00) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y84(1.27) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y84(1.26) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y84(1.74) | LDD0268 | [4] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [6] |
| LDCM0123 | JWB131 | DM93 | Y41(0.82); Y43(1.32); Y84(1.51) | LDD0285 | [3] |
| LDCM0124 | JWB142 | DM93 | Y41(1.06); Y43(0.61); Y84(1.16) | LDD0286 | [3] |
| LDCM0125 | JWB146 | DM93 | Y38(0.39); Y41(0.42); Y43(0.85); Y84(2.41) | LDD0287 | [3] |
| LDCM0126 | JWB150 | DM93 | Y38(1.12); Y41(6.70); Y43(3.81); Y84(6.19) | LDD0288 | [3] |
| LDCM0127 | JWB152 | DM93 | Y41(3.20); Y43(3.12); Y84(2.74) | LDD0289 | [3] |
| LDCM0128 | JWB198 | DM93 | Y41(0.36); Y43(0.82); Y84(4.00) | LDD0290 | [3] |
| LDCM0129 | JWB202 | DM93 | Y41(0.55); Y43(0.37); Y84(1.54) | LDD0291 | [3] |
| LDCM0130 | JWB211 | DM93 | Y38(0.20); Y41(0.57); Y43(0.72); Y84(2.02) | LDD0292 | [3] |
The Interaction Atlas With This Target
References
