Details of the Target
General Information of Target
| Target ID | LDTP14411 | |||||
|---|---|---|---|---|---|---|
| Target Name | Nascent polypeptide-associated complex subunit alpha, muscle-specific form (NACA) | |||||
| Gene Name | NACA | |||||
| Synonyms |
Nascent polypeptide-associated complex subunit alpha, muscle-specific form; Alpha-NAC, muscle-specific form; skNAC |
|||||
| 3D Structure | ||||||
| Sequence |
MASRGALRRCLSPGLPRLLHLSRGLA
|
|||||
| Target Bioclass |
Transcription factor
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Cardiac- and muscle-specific transcription factor. May act to regulate the expression of genes involved in the development of myotubes. Plays a critical role in ventricular cardiomyocyte expansion and regulates postnatal skeletal muscle growth and regeneration. Involved in the organized assembly of thick and thin filaments of myofibril sarcomeres.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K1945(7.75); K1963(10.00); K1971(10.00); K2005(10.00) | LDD0277 | [1] | |
|
AZ-9 Probe Info |
 |
E2004(1.00); E2045(0.86); E1988(0.95) | LDD2208 | [2] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [1] | |
|
P11 Probe Info |
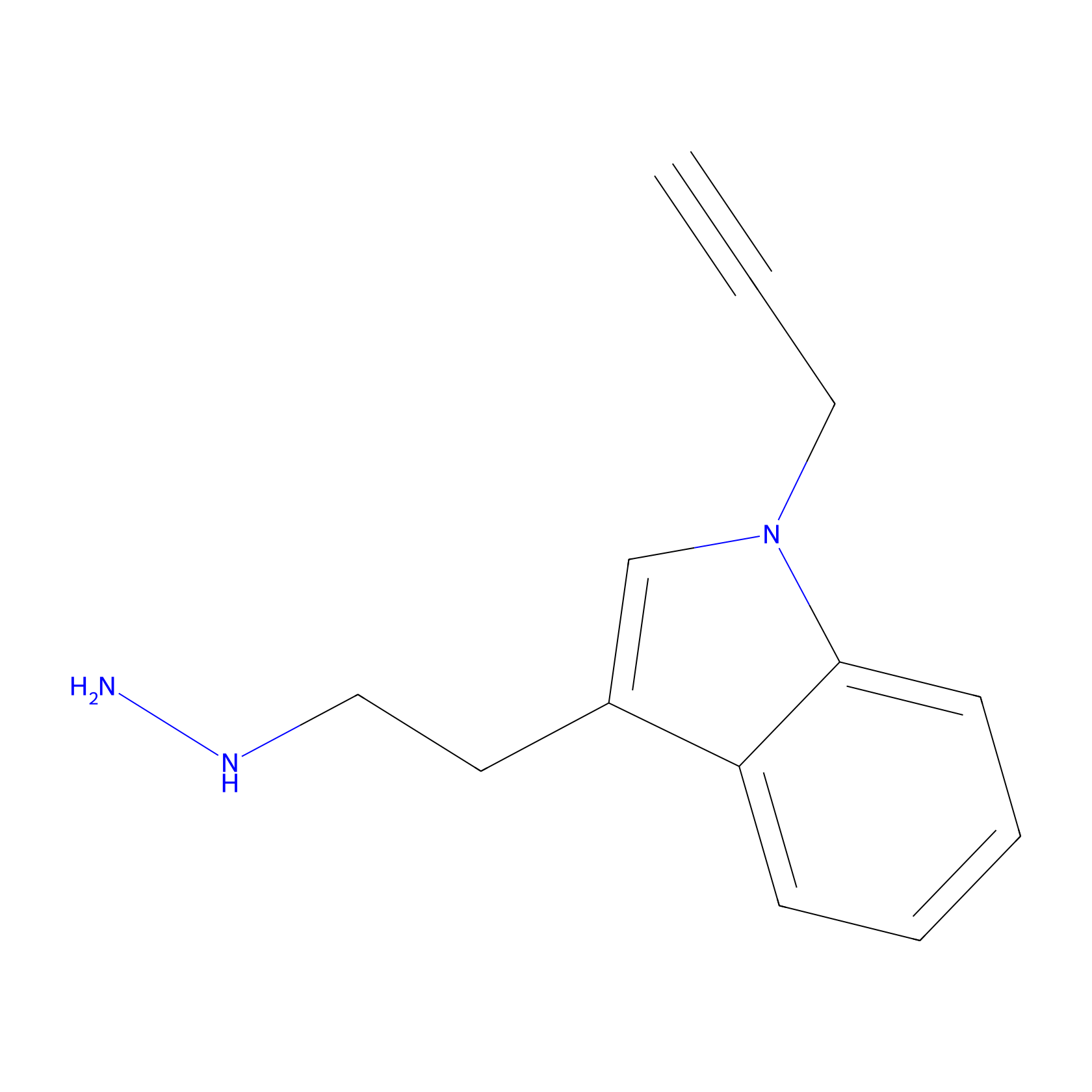 |
5.69 | LDD0201 | [3] | |
|
DBIA Probe Info |
 |
C1717(2.42) | LDD3401 | [4] | |
|
HHS-482 Probe Info |
 |
Y1975(0.09) | LDD0287 | [5] | |
|
HHS-475 Probe Info |
 |
Y1975(0.84); Y1983(0.94) | LDD0264 | [6] | |
|
HHS-465 Probe Info |
 |
Y1983(10.00) | LDD2237 | [7] | |
|
1c-yne Probe Info |
 |
K1963(0.00); K1971(0.00); K1976(0.00); K1990(0.00) | LDD0228 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Photonaproxen Probe Info |
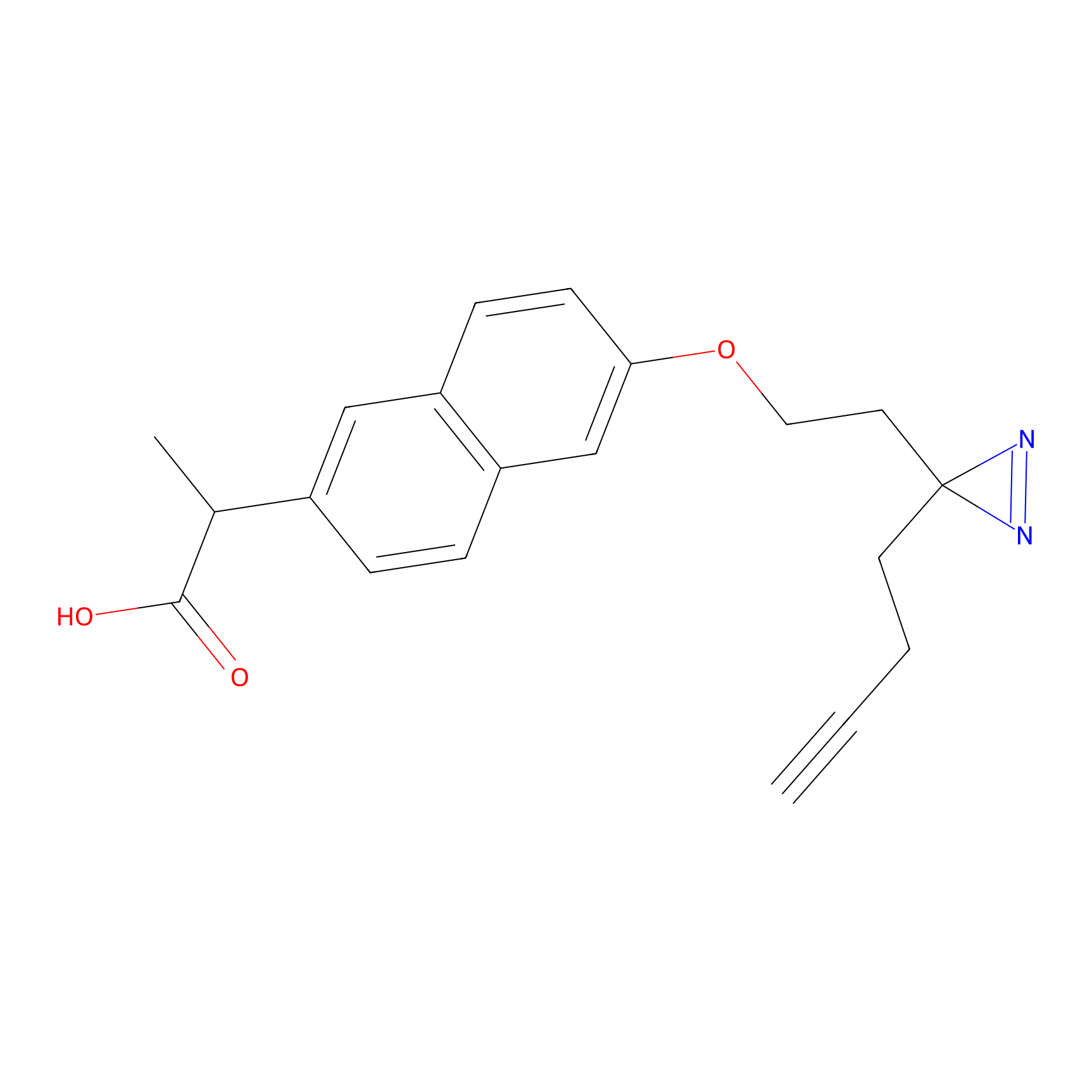 |
I1991(0.00); L1994(0.00) | LDD0156 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0088 | C45 | HEK-293T | 5.69 | LDD0201 | [3] |
| LDCM0116 | HHS-0101 | DM93 | Y1975(0.84); Y1983(0.94) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y1975(0.71); Y1983(1.06) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y1975(0.90); Y1983(1.32) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y1975(0.84); Y1983(1.29) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y1975(0.74); Y1983(0.98) | LDD0268 | [6] |
| LDCM0125 | JWB146 | DM93 | Y1975(0.09) | LDD0287 | [5] |
| LDCM0126 | JWB150 | DM93 | Y1975(0.14) | LDD0288 | [5] |
| LDCM0127 | JWB152 | DM93 | Y1975(0.26) | LDD0289 | [5] |
| LDCM0128 | JWB198 | DM93 | Y1975(0.07) | LDD0290 | [5] |
| LDCM0129 | JWB202 | DM93 | Y1975(0.09) | LDD0291 | [5] |
| LDCM0130 | JWB211 | DM93 | Y1975(0.18) | LDD0292 | [5] |
| LDCM0022 | KB02 | RD | C1717(1.77) | LDD2567 | [4] |
| LDCM0023 | KB03 | RD | C1717(1.66) | LDD2984 | [4] |
| LDCM0024 | KB05 | RD | C1717(2.42) | LDD3401 | [4] |
References
