Details of the Target
General Information of Target
| Target ID | LDTP11657 | |||||
|---|---|---|---|---|---|---|
| Target Name | Eukaryotic translation initiation factor 5A-2 (EIF5A2) | |||||
| Gene Name | EIF5A2 | |||||
| Gene ID | 56648 | |||||
| Synonyms |
Eukaryotic translation initiation factor 5A-2; eIF-5A-2; eIF-5A2; Eukaryotic initiation factor 5A isoform 2 |
|||||
| 3D Structure | ||||||
| Sequence |
MAFHVEGLIAIIVFYLLILLVGIWAAWRTKNSGSAEERSEAIIVGGRDIGLLVGGFTMTA
TWVGGGYINGTAEAVYVPGYGLAWAQAPIGYSLSLILGGLFFAKPMRSKGYVTMLDPFQQ IYGKRMGGLLFIPALMGEMFWAAAIFSALGATISVIIDVDMHISVIISALIATLYTLVGG LYSVAYTDVVQLFCIFVGLWISVPFALSHPAVADIGFTAVHAKYQKPWLGTVDSSEVYSW LDSFLLLMLGGIPWQAYFQRVLSSSSATYAQVLSFLAAFGCLVMAIPAILIGAIGASTDW NQTAYGLPDPKTTEEADMILPIVLQYLCPVYISFFGLGAVSAAVMSSADSSILSASSMFA RNIYQLSFRQNASDKEIVWVMRITVFVFGASATAMALLTKTVYGLWYLSSDLVYIVIFPQ LLCVLFVKGTNTYGAVAGYVSGLFLRITGGEPYLYLQPLIFYPGYYPDDNGIYNQKFPFK TLAMVTSFLTNICISYLAKYLFESGTLPPKLDVFDAVVARHSEENMDKTILVKNENIKLD ELALVKPRQSMTLSSTFTNKEAFLDVDSSPEGSGTEDNLQ |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
EIF-5A family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Translation factor that promotes translation elongation and termination, particularly upon ribosome stalling at specific amino acid sequence contexts. Binds between the exit (E) and peptidyl (P) site of the ribosome and promotes rescue of stalled ribosome: specifically required for efficient translation of polyproline-containing peptides as well as other motifs that stall the ribosome. Acts as a ribosome quality control (RQC) cofactor by joining the RQC complex to facilitate peptidyl transfer during CAT tailing step. Also involved in actin dynamics and cell cycle progression, mRNA decay and probably in a pathway involved in stress response and maintenance of cell wall integrity.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
AZ-9 Probe Info |
 |
E42(0.86) | LDD2208 | [2] | |
|
DBIA Probe Info |
 |
C73(0.83); C93(1.40) | LDD3331 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C93(3.85) | LDD0170 | [4] | |
|
HHS-475 Probe Info |
 |
Y69(0.89) | LDD0264 | [5] | |
|
HHS-465 Probe Info |
 |
Y69(10.00) | LDD2237 | [6] | |
|
W1 Probe Info |
 |
C73(0.87) | LDD0237 | [7] | |
|
IA-alkyne Probe Info |
 |
C93(0.00); C73(0.00) | LDD0174 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [9] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2225 | [10] | |
|
aHNE Probe Info |
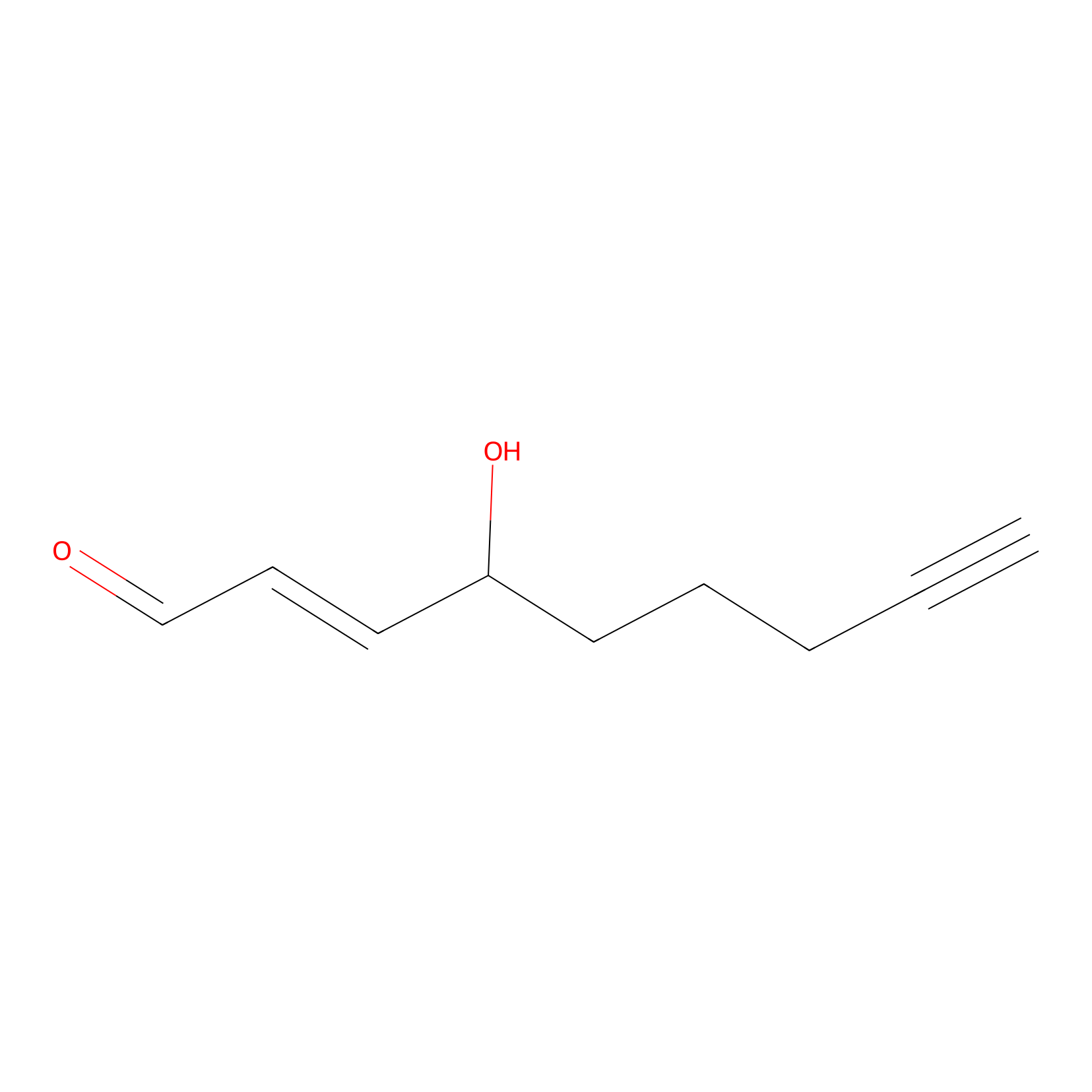 |
N.A. | LDD0001 | [11] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [12] | |
|
SF Probe Info |
 |
K27(0.00); K67(0.00); K68(0.00); K39(0.00) | LDD0028 | [13] | |
|
STPyne Probe Info |
 |
K68(0.00); K39(0.00); K67(0.00) | LDD0009 | [11] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe13 Probe Info |
 |
6.21 | LDD0475 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C93(3.85) | LDD0170 | [4] |
| LDCM0226 | AC11 | HEK-293T | C93(1.03) | LDD1509 | [16] |
| LDCM0230 | AC113 | PaTu 8988t | C139(0.92); C152(0.92) | LDD1109 | [17] |
| LDCM0231 | AC114 | PaTu 8988t | C139(0.60); C152(0.60) | LDD1110 | [17] |
| LDCM0232 | AC115 | PaTu 8988t | C139(0.81); C152(0.81) | LDD1111 | [17] |
| LDCM0233 | AC116 | PaTu 8988t | C139(0.91); C152(0.91) | LDD1112 | [17] |
| LDCM0234 | AC117 | PaTu 8988t | C139(0.43); C152(0.43) | LDD1113 | [17] |
| LDCM0235 | AC118 | PaTu 8988t | C139(0.94); C152(0.94) | LDD1114 | [17] |
| LDCM0236 | AC119 | PaTu 8988t | C139(0.88); C152(0.88) | LDD1115 | [17] |
| LDCM0238 | AC120 | PaTu 8988t | C139(1.09); C152(1.09) | LDD1117 | [17] |
| LDCM0239 | AC121 | PaTu 8988t | C139(0.51); C152(0.51) | LDD1118 | [17] |
| LDCM0240 | AC122 | PaTu 8988t | C139(1.08); C152(1.08) | LDD1119 | [17] |
| LDCM0241 | AC123 | PaTu 8988t | C139(1.55); C152(1.55) | LDD1120 | [17] |
| LDCM0242 | AC124 | PaTu 8988t | C139(1.82); C152(1.82) | LDD1121 | [17] |
| LDCM0243 | AC125 | PaTu 8988t | C139(2.31); C152(2.31) | LDD1122 | [17] |
| LDCM0244 | AC126 | PaTu 8988t | C139(1.11); C152(1.11) | LDD1123 | [17] |
| LDCM0245 | AC127 | PaTu 8988t | C139(1.01); C152(1.01) | LDD1124 | [17] |
| LDCM0246 | AC128 | PaTu 8988t | C139(0.43); C152(0.43) | LDD1125 | [17] |
| LDCM0247 | AC129 | PaTu 8988t | C139(0.62); C152(0.62) | LDD1126 | [17] |
| LDCM0249 | AC130 | PaTu 8988t | C139(0.55); C152(0.55) | LDD1128 | [17] |
| LDCM0250 | AC131 | PaTu 8988t | C139(0.88); C152(0.88) | LDD1129 | [17] |
| LDCM0251 | AC132 | PaTu 8988t | C139(0.99); C152(0.99) | LDD1130 | [17] |
| LDCM0252 | AC133 | PaTu 8988t | C139(0.75); C152(0.75) | LDD1131 | [17] |
| LDCM0253 | AC134 | PaTu 8988t | C139(0.67); C152(0.67) | LDD1132 | [17] |
| LDCM0254 | AC135 | PaTu 8988t | C139(0.59); C152(0.59) | LDD1133 | [17] |
| LDCM0255 | AC136 | PaTu 8988t | C139(0.32); C152(0.32) | LDD1134 | [17] |
| LDCM0256 | AC137 | PaTu 8988t | C139(0.51); C152(0.51) | LDD1135 | [17] |
| LDCM0257 | AC138 | PaTu 8988t | C139(0.45); C152(0.45) | LDD1136 | [17] |
| LDCM0258 | AC139 | PaTu 8988t | C139(0.42); C152(0.42) | LDD1137 | [17] |
| LDCM0260 | AC140 | PaTu 8988t | C139(0.82); C152(0.82) | LDD1139 | [17] |
| LDCM0261 | AC141 | PaTu 8988t | C139(1.02); C152(1.02) | LDD1140 | [17] |
| LDCM0262 | AC142 | PaTu 8988t | C139(0.74); C152(0.74) | LDD1141 | [17] |
| LDCM0270 | AC15 | HEK-293T | C93(1.13) | LDD1513 | [16] |
| LDCM0278 | AC19 | HEK-293T | C93(1.17) | LDD1517 | [16] |
| LDCM0283 | AC23 | HEK-293T | C93(1.01) | LDD1522 | [16] |
| LDCM0287 | AC27 | HEK-293T | C93(1.02) | LDD1526 | [16] |
| LDCM0290 | AC3 | HEK-293T | C93(1.00) | LDD1529 | [16] |
| LDCM0292 | AC31 | HEK-293T | C93(0.96) | LDD1531 | [16] |
| LDCM0296 | AC35 | HEK-293T | C93(0.94) | LDD1535 | [16] |
| LDCM0300 | AC39 | HEK-293T | C93(1.36) | LDD1539 | [16] |
| LDCM0305 | AC43 | HEK-293T | C93(1.30) | LDD1544 | [16] |
| LDCM0308 | AC46 | PaTu 8988t | C139(0.96); C152(0.96) | LDD1187 | [17] |
| LDCM0309 | AC47 | PaTu 8988t | C139(0.92); C152(0.92) | LDD1188 | [17] |
| LDCM0310 | AC48 | PaTu 8988t | C139(1.22); C152(1.22) | LDD1189 | [17] |
| LDCM0311 | AC49 | PaTu 8988t | C139(1.35); C152(1.35) | LDD1190 | [17] |
| LDCM0313 | AC50 | PaTu 8988t | C139(0.85); C152(0.85) | LDD1192 | [17] |
| LDCM0314 | AC51 | PaTu 8988t | C139(0.88); C152(0.88) | LDD1193 | [17] |
| LDCM0315 | AC52 | PaTu 8988t | C139(1.17); C152(1.17) | LDD1194 | [17] |
| LDCM0316 | AC53 | PaTu 8988t | C139(1.68); C152(1.68) | LDD1195 | [17] |
| LDCM0317 | AC54 | PaTu 8988t | C139(2.26); C152(2.26) | LDD1196 | [17] |
| LDCM0318 | AC55 | PaTu 8988t | C139(1.17); C152(1.17) | LDD1197 | [17] |
| LDCM0319 | AC56 | PaTu 8988t | C139(0.99); C152(0.99) | LDD1198 | [17] |
| LDCM0322 | AC59 | HEK-293T | C93(1.02) | LDD1561 | [16] |
| LDCM0327 | AC63 | HEK-293T | C93(0.81) | LDD1566 | [16] |
| LDCM0332 | AC68 | PaTu 8988t | C139(0.86); C152(0.86) | LDD1211 | [17] |
| LDCM0333 | AC69 | PaTu 8988t | C139(0.90); C152(0.90) | LDD1212 | [17] |
| LDCM0334 | AC7 | HEK-293T | C93(0.87) | LDD1568 | [16] |
| LDCM0335 | AC70 | PaTu 8988t | C139(0.63); C152(0.63) | LDD1214 | [17] |
| LDCM0336 | AC71 | PaTu 8988t | C139(0.81); C152(0.81) | LDD1215 | [17] |
| LDCM0337 | AC72 | PaTu 8988t | C139(0.84); C152(0.84) | LDD1216 | [17] |
| LDCM0338 | AC73 | PaTu 8988t | C139(0.84); C152(0.84) | LDD1217 | [17] |
| LDCM0339 | AC74 | PaTu 8988t | C139(0.93); C152(0.93) | LDD1218 | [17] |
| LDCM0340 | AC75 | PaTu 8988t | C139(0.79); C152(0.79) | LDD1219 | [17] |
| LDCM0341 | AC76 | PaTu 8988t | C139(1.57); C152(1.57) | LDD1220 | [17] |
| LDCM0342 | AC77 | PaTu 8988t | C139(0.84); C152(0.84) | LDD1221 | [17] |
| LDCM0343 | AC78 | PaTu 8988t | C139(1.02); C152(1.02) | LDD1222 | [17] |
| LDCM0344 | AC79 | PaTu 8988t | C139(0.70); C152(0.70) | LDD1223 | [17] |
| LDCM0346 | AC80 | PaTu 8988t | C139(0.65); C152(0.65) | LDD1225 | [17] |
| LDCM0347 | AC81 | PaTu 8988t | C139(0.80); C152(0.80) | LDD1226 | [17] |
| LDCM0348 | AC82 | PaTu 8988t | C139(1.47); C152(1.47) | LDD1227 | [17] |
| LDCM0156 | Aniline | NCI-H1299 | 13.85 | LDD0403 | [1] |
| LDCM0367 | CL1 | PaTu 8988t | C139(1.17); C152(1.17) | LDD1246 | [17] |
| LDCM0368 | CL10 | PaTu 8988t | C139(1.63); C152(1.63) | LDD1247 | [17] |
| LDCM0370 | CL101 | HEK-293T | C93(1.08) | LDD1574 | [16] |
| LDCM0371 | CL102 | HEK-293T | C93(0.89) | LDD1575 | [16] |
| LDCM0374 | CL105 | HEK-293T | C93(0.97) | LDD1578 | [16] |
| LDCM0375 | CL106 | HEK-293T | C93(1.29) | LDD1579 | [16] |
| LDCM0378 | CL109 | HEK-293T | C93(1.37) | LDD1582 | [16] |
| LDCM0379 | CL11 | PaTu 8988t | C139(1.10); C152(1.10) | LDD1258 | [17] |
| LDCM0380 | CL110 | HEK-293T | C93(1.12) | LDD1584 | [16] |
| LDCM0383 | CL113 | HEK-293T | C93(1.66) | LDD1587 | [16] |
| LDCM0384 | CL114 | HEK-293T | C93(1.01) | LDD1588 | [16] |
| LDCM0387 | CL117 | HEK-293T | C93(1.06) | LDD1591 | [16] |
| LDCM0388 | CL118 | HEK-293T | C93(1.18) | LDD1592 | [16] |
| LDCM0390 | CL12 | PaTu 8988t | C139(2.20); C152(2.20) | LDD1269 | [17] |
| LDCM0392 | CL121 | PaTu 8988t | C139(0.90); C152(0.90) | LDD1271 | [17] |
| LDCM0393 | CL122 | PaTu 8988t | C139(1.65); C152(1.65) | LDD1272 | [17] |
| LDCM0394 | CL123 | PaTu 8988t | C139(1.35); C152(1.35) | LDD1273 | [17] |
| LDCM0395 | CL124 | PaTu 8988t | C139(1.58); C152(1.58) | LDD1274 | [17] |
| LDCM0396 | CL125 | HEK-293T | C93(0.74) | LDD1600 | [16] |
| LDCM0397 | CL126 | HEK-293T | C93(1.13) | LDD1601 | [16] |
| LDCM0400 | CL13 | PaTu 8988t | C139(2.88); C152(2.88) | LDD1279 | [17] |
| LDCM0401 | CL14 | PaTu 8988t | C139(7.04); C152(7.04) | LDD1280 | [17] |
| LDCM0402 | CL15 | PaTu 8988t | C139(2.19); C152(2.19) | LDD1281 | [17] |
| LDCM0406 | CL19 | HEK-293T | C93(1.25) | LDD1610 | [16] |
| LDCM0407 | CL2 | PaTu 8988t | C139(2.07); C152(2.07) | LDD1286 | [17] |
| LDCM0411 | CL23 | HEK-293T | C93(1.09) | LDD1615 | [16] |
| LDCM0413 | CL25 | HEK-293T | C93(1.36) | LDD1617 | [16] |
| LDCM0414 | CL26 | HEK-293T | C93(1.59) | LDD1618 | [16] |
| LDCM0418 | CL3 | PaTu 8988t | C139(1.22); C152(1.22) | LDD1297 | [17] |
| LDCM0420 | CL31 | HEK-293T | C93(0.95) | LDD1624 | [16] |
| LDCM0424 | CL35 | HEK-293T | C93(1.93) | LDD1628 | [16] |
| LDCM0426 | CL37 | HEK-293T | C93(0.44) | LDD1630 | [16] |
| LDCM0429 | CL4 | PaTu 8988t | C139(3.09); C152(3.09) | LDD1308 | [17] |
| LDCM0433 | CL43 | HEK-293T | C93(1.03) | LDD1637 | [16] |
| LDCM0437 | CL47 | HEK-293T | C93(0.91) | LDD1641 | [16] |
| LDCM0439 | CL49 | HEK-293T | C93(1.13) | LDD1643 | [16] |
| LDCM0440 | CL5 | PaTu 8988t | C139(2.59); C152(2.59) | LDD1319 | [17] |
| LDCM0441 | CL50 | HEK-293T | C93(1.39) | LDD1645 | [16] |
| LDCM0446 | CL55 | HEK-293T | C93(1.19) | LDD1649 | [16] |
| LDCM0450 | CL59 | HEK-293T | C93(1.28) | LDD1653 | [16] |
| LDCM0451 | CL6 | PaTu 8988t | C139(3.01); C152(3.01) | LDD1330 | [17] |
| LDCM0453 | CL61 | HEK-293T | C93(1.44) | LDD1656 | [16] |
| LDCM0454 | CL62 | HEK-293T | C93(0.98) | LDD1657 | [16] |
| LDCM0459 | CL67 | HEK-293T | C93(1.25) | LDD1662 | [16] |
| LDCM0462 | CL7 | PaTu 8988t | C139(1.63); C152(1.63) | LDD1341 | [17] |
| LDCM0464 | CL71 | HEK-293T | C93(0.88) | LDD1667 | [16] |
| LDCM0466 | CL73 | HEK-293T | C93(1.10) | LDD1669 | [16] |
| LDCM0467 | CL74 | HEK-293T | C93(1.36) | LDD1670 | [16] |
| LDCM0469 | CL76 | PaTu 8988t | C139(0.74); C152(0.74) | LDD1348 | [17] |
| LDCM0470 | CL77 | PaTu 8988t | C139(0.79); C152(0.79) | LDD1349 | [17] |
| LDCM0471 | CL78 | PaTu 8988t | C139(0.57); C152(0.57) | LDD1350 | [17] |
| LDCM0472 | CL79 | PaTu 8988t | C139(0.40); C152(0.40) | LDD1351 | [17] |
| LDCM0473 | CL8 | PaTu 8988t | C139(1.37); C152(1.37) | LDD1352 | [17] |
| LDCM0474 | CL80 | PaTu 8988t | C139(0.91); C152(0.91) | LDD1353 | [17] |
| LDCM0475 | CL81 | PaTu 8988t | C139(1.17); C152(1.17) | LDD1354 | [17] |
| LDCM0476 | CL82 | PaTu 8988t | C139(0.60); C152(0.60) | LDD1355 | [17] |
| LDCM0477 | CL83 | PaTu 8988t | C139(0.86); C152(0.86) | LDD1356 | [17] |
| LDCM0478 | CL84 | PaTu 8988t | C139(0.98); C152(0.98) | LDD1357 | [17] |
| LDCM0479 | CL85 | PaTu 8988t | C139(0.87); C152(0.87) | LDD1358 | [17] |
| LDCM0480 | CL86 | PaTu 8988t | C139(1.30); C152(1.30) | LDD1359 | [17] |
| LDCM0481 | CL87 | PaTu 8988t | C139(1.79); C152(1.79) | LDD1360 | [17] |
| LDCM0482 | CL88 | PaTu 8988t | C139(1.73); C152(1.73) | LDD1361 | [17] |
| LDCM0483 | CL89 | PaTu 8988t | C139(1.56); C152(1.56) | LDD1362 | [17] |
| LDCM0484 | CL9 | PaTu 8988t | C139(3.09); C152(3.09) | LDD1363 | [17] |
| LDCM0485 | CL90 | PaTu 8988t | C139(1.76); C152(1.76) | LDD1364 | [17] |
| LDCM0486 | CL91 | HEK-293T | C93(1.01) | LDD1689 | [16] |
| LDCM0490 | CL95 | HEK-293T | C93(0.98) | LDD1693 | [16] |
| LDCM0492 | CL97 | HEK-293T | C93(1.26) | LDD1695 | [16] |
| LDCM0493 | CL98 | HEK-293T | C93(0.85) | LDD1696 | [16] |
| LDCM0625 | F8 | Ramos | C73(1.32) | LDD2187 | [18] |
| LDCM0572 | Fragment10 | Ramos | C73(0.42) | LDD2189 | [18] |
| LDCM0573 | Fragment11 | Ramos | C73(0.01) | LDD2190 | [18] |
| LDCM0574 | Fragment12 | Ramos | C73(0.50) | LDD2191 | [18] |
| LDCM0575 | Fragment13 | Ramos | C73(1.18) | LDD2192 | [18] |
| LDCM0576 | Fragment14 | Ramos | C73(0.57) | LDD2193 | [18] |
| LDCM0579 | Fragment20 | Ramos | C73(0.38) | LDD2194 | [18] |
| LDCM0580 | Fragment21 | Ramos | C73(0.91) | LDD2195 | [18] |
| LDCM0582 | Fragment23 | Ramos | C73(0.37) | LDD2196 | [18] |
| LDCM0578 | Fragment27 | Ramos | C73(1.40) | LDD2197 | [18] |
| LDCM0586 | Fragment28 | Ramos | C73(0.68) | LDD2198 | [18] |
| LDCM0588 | Fragment30 | Ramos | C73(0.99) | LDD2199 | [18] |
| LDCM0589 | Fragment31 | Ramos | C73(1.02) | LDD2200 | [18] |
| LDCM0590 | Fragment32 | Ramos | C73(0.42) | LDD2201 | [18] |
| LDCM0468 | Fragment33 | Ramos | C73(0.95) | LDD2202 | [18] |
| LDCM0596 | Fragment38 | Ramos | C73(1.49) | LDD2203 | [18] |
| LDCM0566 | Fragment4 | Ramos | C73(0.50) | LDD2184 | [18] |
| LDCM0427 | Fragment51 | HEK-293T | C93(2.20) | LDD1631 | [16] |
| LDCM0610 | Fragment52 | Ramos | C73(1.12) | LDD2204 | [18] |
| LDCM0614 | Fragment56 | Ramos | C73(1.03) | LDD2205 | [18] |
| LDCM0569 | Fragment7 | Ramos | C73(0.45) | LDD2186 | [18] |
| LDCM0571 | Fragment9 | Ramos | C73(0.26) | LDD2188 | [18] |
| LDCM0116 | HHS-0101 | DM93 | Y69(0.89) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y69(0.71) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y69(0.68) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y69(0.77) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y69(0.97) | LDD0268 | [5] |
| LDCM0022 | KB02 | HEK-293T | C93(0.87) | LDD1492 | [16] |
| LDCM0023 | KB03 | HEK-293T | C93(1.12) | LDD1497 | [16] |
| LDCM0024 | KB05 | MKN-1 | C73(0.83); C93(1.40) | LDD3331 | [3] |
| LDCM0110 | W12 | Hep-G2 | C73(0.87) | LDD0237 | [7] |
| LDCM0111 | W14 | Hep-G2 | C73(0.65) | LDD0238 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Syntenin-1 (SDCBP) | . | O00560 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Proto-oncogene c-Rel (REL) | . | Q04864 | |||
References
