Details of the Target
General Information of Target
| Target ID | LDTP09562 | |||||
|---|---|---|---|---|---|---|
| Target Name | Splicing factor U2AF 26 kDa subunit (U2AF1L4) | |||||
| Gene Name | U2AF1L4 | |||||
| Gene ID | 199746 | |||||
| Synonyms |
U2AF1-RS3; U2AF1L3; Splicing factor U2AF 26 kDa subunit; U2 auxiliary factor 26; U2 small nuclear RNA auxiliary factor 1-like protein 4; U2AF1-like 4; U2(RNU2) small nuclear RNA auxiliary factor 1-like protein 3; U2 small nuclear RNA auxiliary factor 1-like protein 3; U2AF1-like protein 3
|
|||||
| 3D Structure | ||||||
| Sequence |
MAEYLASIFGTEKDKVNCSFYFKIGVCRHGDRCSRLHNKPTFSQTIVLLNLYRNPQNTAQ
TADGSHCHVSDVEVQEHYDSFFEEVFTELQEKYGEIEEMNVCDNLGDHLVGNVYVKFRRE EDGERAVAELSNRWFNGQAVHGELSPVTDFRESCCRQYEMGECTRGGFCNFMHLRPISQN LQRQLYGRGPRRRSPPRFHTGHHPRERNHRCSPDHWHGRF |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Splicing factor SR family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
RNA-binding protein that function as a pre-mRNA splicing factor. Plays a critical role in both constitutive and enhancer-dependent splicing by mediating protein-protein interactions and protein-RNA interactions required for accurate 3'-splice site selection. Acts by enhancing the binding of U2AF2 to weak pyrimidine tracts. Also participates in the regulation of alternative pre-mRNA splicing. Activates exon 5 skipping of PTPRC during T-cell activation; an event reversed by GFI1. Binds to RNA at the AG dinucleotide at the 3'-splice site. Shows a preference for AGC or AGA.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P1 Probe Info |
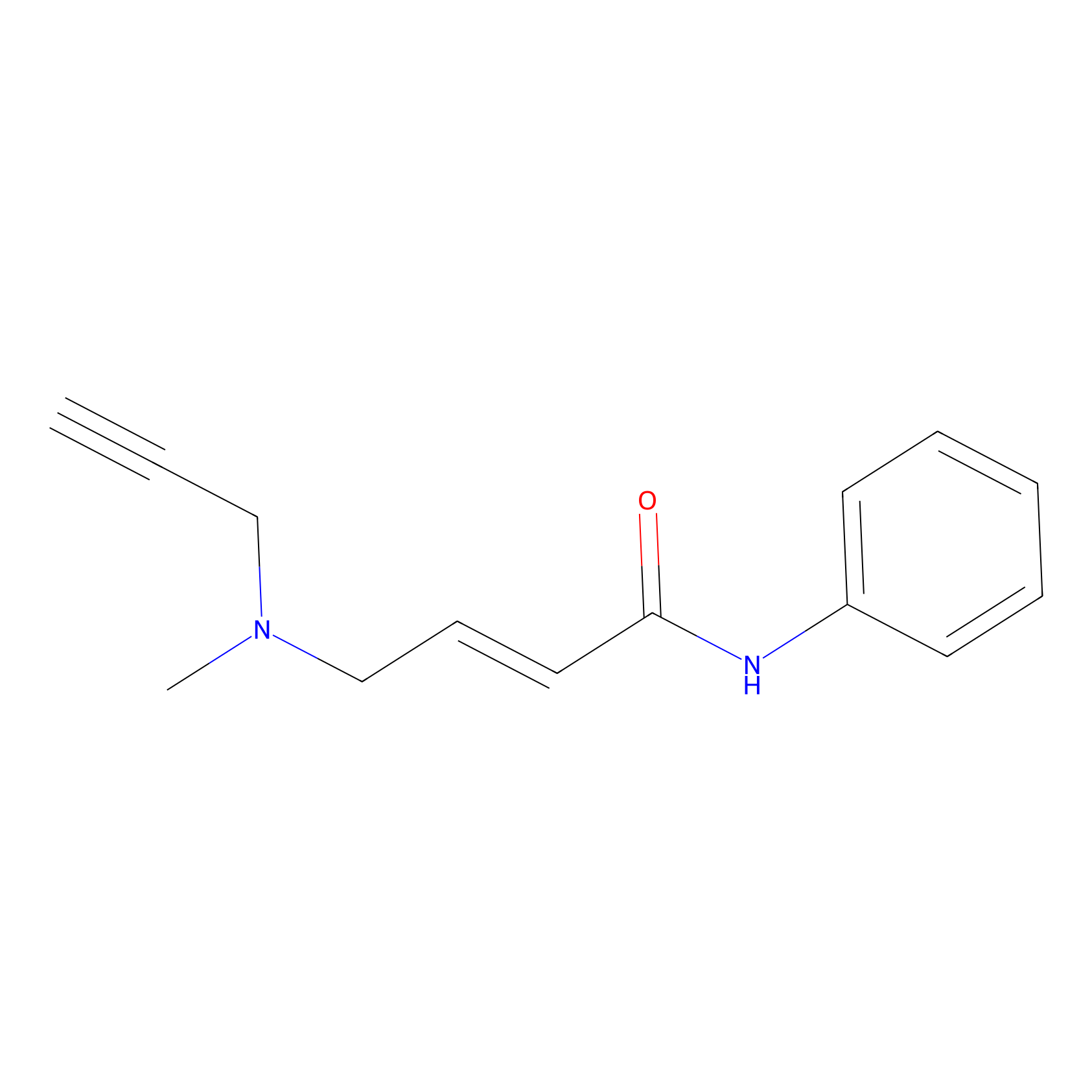 |
2.79 | LDD0452 | [1] | |
|
P3 Probe Info |
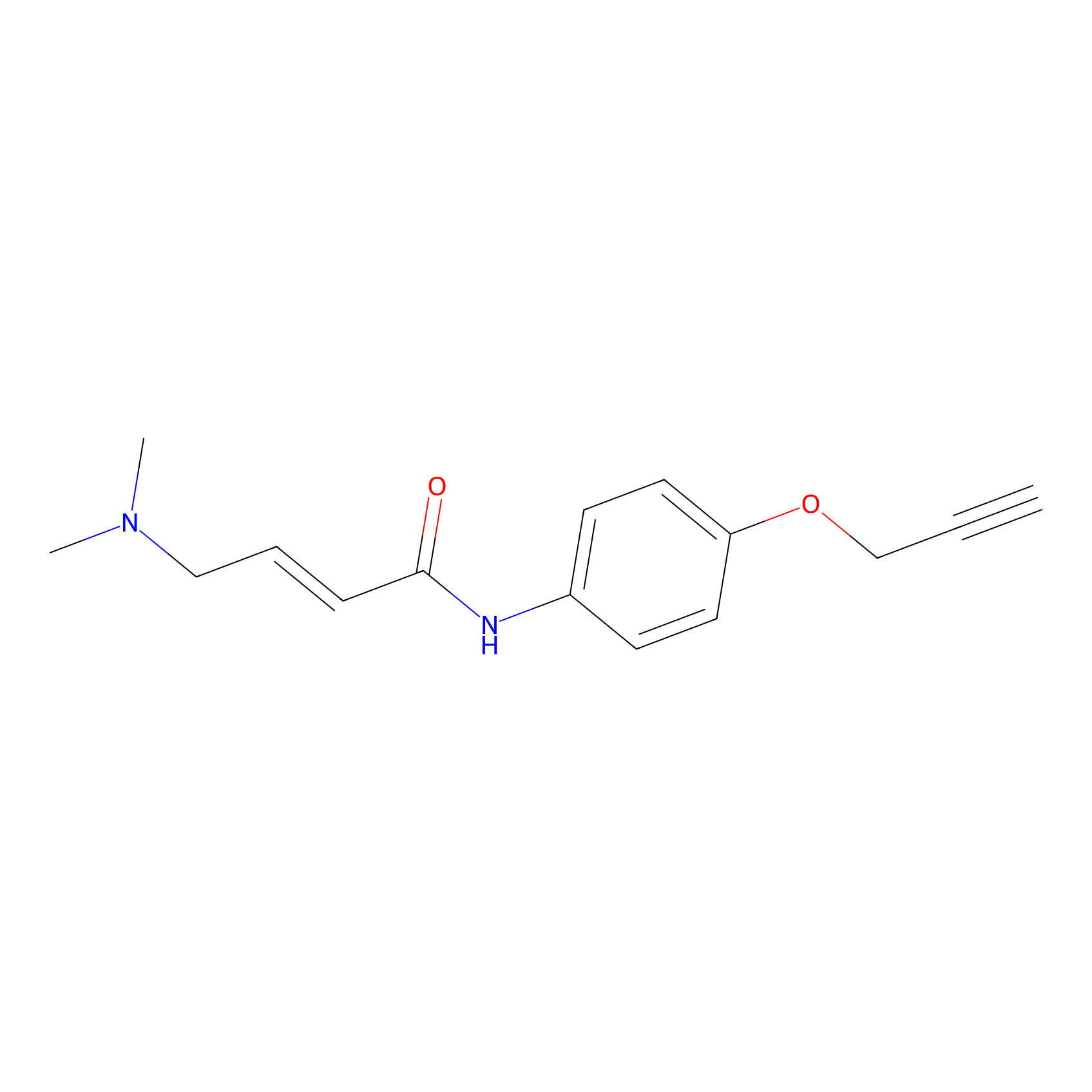 |
1.76 | LDD0454 | [1] | |
|
P8 Probe Info |
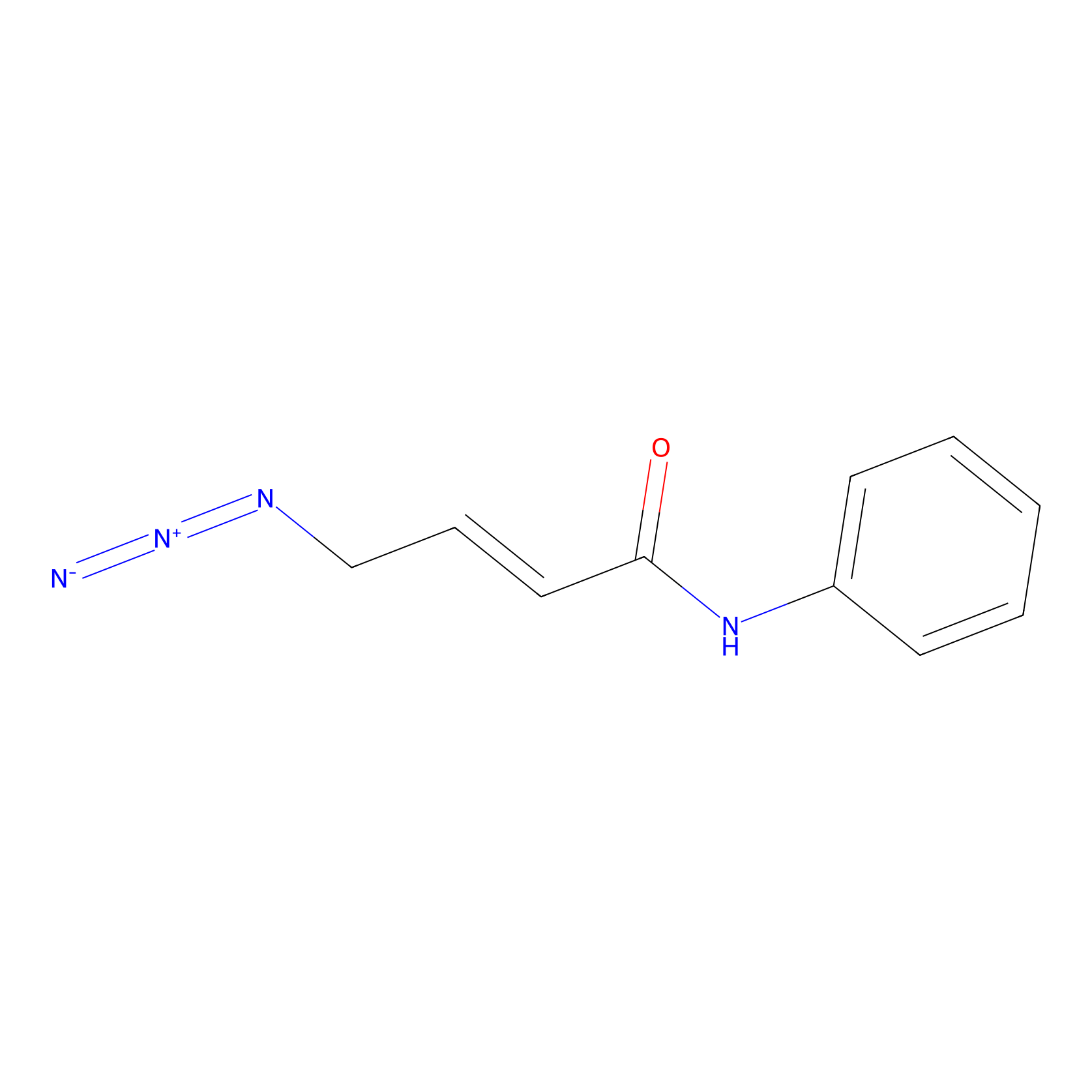 |
6.26 | LDD0455 | [1] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [2] | |
|
DBIA Probe Info |
 |
C90(1.21) | LDD3310 | [3] | |
|
IA-alkyne Probe Info |
 |
C163(10.21) | LDD0372 | [4] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [6] | |
Competitor(s) Related to This Target
References
