Details of the Target
General Information of Target
| Target ID | LDTP05853 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ubiquitin-conjugating enzyme E2 variant 1 (UBE2V1) | |||||
| Gene Name | UBE2V1 | |||||
| Gene ID | 387522 | |||||
| Synonyms |
CROC1; UBE2V; UEV1; Ubiquitin-conjugating enzyme E2 variant 1; UEV-1; CROC-1; TRAF6-regulated IKK activator 1 beta Uev1A |
|||||
| 3D Structure | ||||||
| Sequence |
MAATTGSGVKVPRNFRLLEELEEGQKGVGDGTVSWGLEDDEDMTLTRWTGMIIGPPRTIY
ENRIYSLKIECGPKYPEAPPFVRFVTKINMNGVNSSNGVVDPRAISVLAKWQNSYSIKVV LQELRRLMMSKENMKLPQPPEGQCYSN |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Ubiquitin-conjugating enzyme family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Has no ubiquitin ligase activity on its own. The UBE2V1-UBE2N heterodimer catalyzes the synthesis of non-canonical poly-ubiquitin chains that are linked through Lys-63. This type of poly-ubiquitination activates IKK and does not seem to involve protein degradation by the proteasome. Plays a role in the activation of NF-kappa-B mediated by IL1B, TNF, TRAF6 and TRAF2. Mediates transcriptional activation of target genes. Plays a role in the control of progress through the cell cycle and differentiation. Plays a role in the error-free DNA repair pathway and contributes to the survival of cells after DNA damage. Promotes TRIM5 capsid-specific restriction activity and the UBE2V1-UBE2N heterodimer acts in concert with TRIM5 to generate 'Lys-63'-linked polyubiquitin chains which activate the MAP3K7/TAK1 complex which in turn results in the induction and expression of NF-kappa-B and MAPK-responsive inflammatory genes. Together with RNF135 and UBE2N, catalyzes the viral RNA-dependent 'Lys-63'-linked polyubiquitination of RIGI to activate the downstream signaling pathway that leads to interferon beta production. UBE2V1-UBE2N together with TRAF3IP2 E3 ubiquitin ligase mediate 'Lys-63'-linked polyubiquitination of TRAF6, a component of IL17A-mediated signaling pathway.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
FBP2 Probe Info |
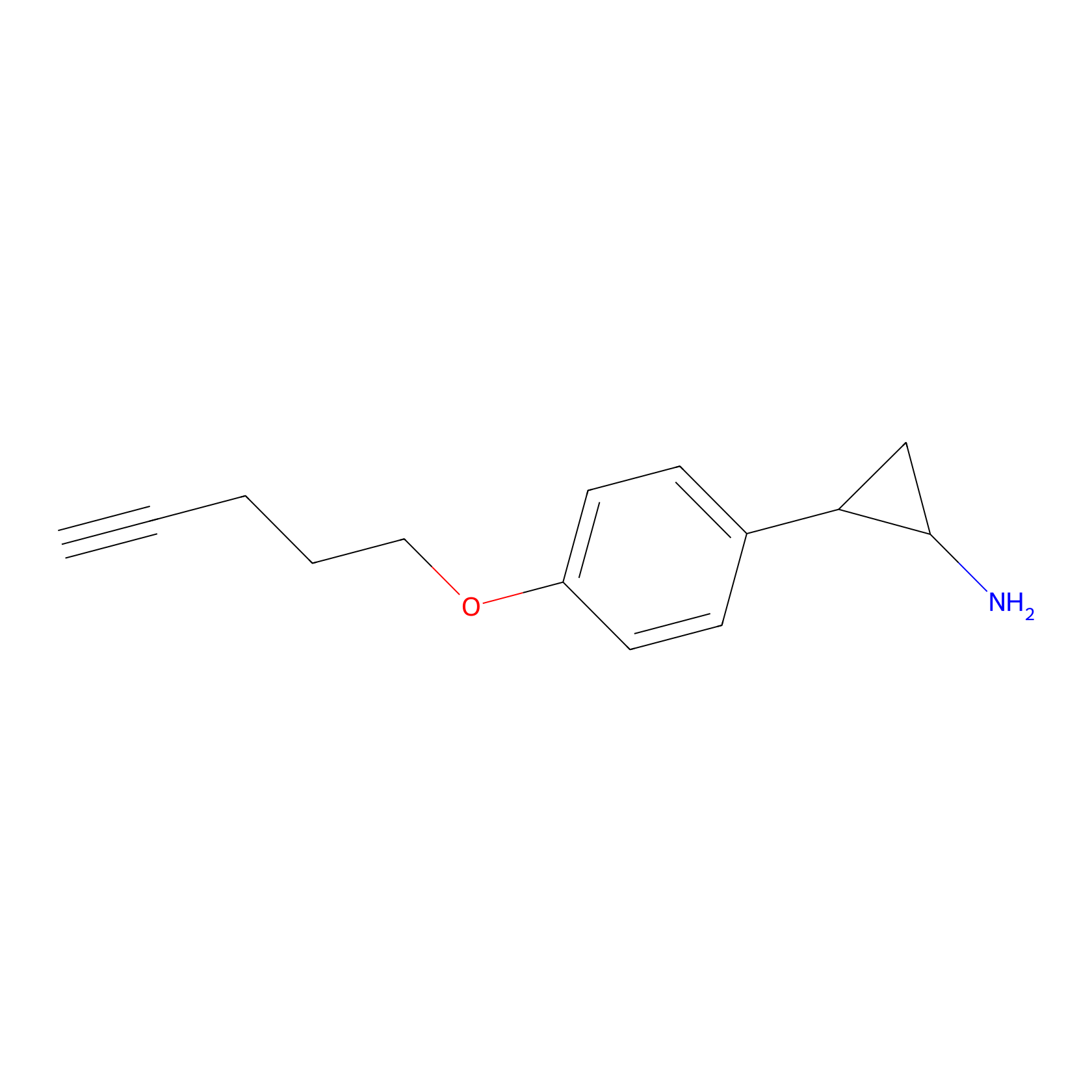 |
2.34 | LDD0323 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
BTD Probe Info |
 |
C71(0.57) | LDD1699 | [4] | |
|
AZ-9 Probe Info |
 |
E22(10.00); E19(10.00) | LDD2209 | [5] | |
|
Probe 1 Probe Info |
 |
Y60(64.44); Y75(11.99); Y115(80.34) | LDD3495 | [6] | |
|
DBIA Probe Info |
 |
C94(2.77); C218(1.55) | LDD3310 | [7] | |
|
HHS-475 Probe Info |
 |
Y115(0.92) | LDD0264 | [8] | |
|
HHS-465 Probe Info |
 |
Y115(10.00) | LDD2237 | [9] | |
|
ATP probe Probe Info |
 |
K74(0.00); K131(0.00) | LDD0199 | [10] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [11] | |
|
IA-alkyne Probe Info |
 |
C144(0.00); C71(0.00) | LDD0036 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [11] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [12] | |
|
TFBX Probe Info |
 |
C71(0.00); C144(0.00) | LDD0027 | [13] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0356 | [14] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [15] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [16] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [17] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [14] | |
|
Acrolein Probe Info |
 |
C71(0.00); C144(0.00) | LDD0217 | [18] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [18] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [19] | |
|
NAIA_5 Probe Info |
 |
C144(0.00); C71(0.00) | LDD2223 | [20] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C71(0.23) | LDD2142 | [4] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C71(0.37) | LDD2112 | [4] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C71(0.47) | LDD2095 | [4] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C71(1.00) | LDD2130 | [4] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C71(0.99) | LDD2117 | [4] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C71(1.23) | LDD2152 | [4] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C71(0.46) | LDD2132 | [4] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C71(0.80) | LDD2131 | [4] |
| LDCM0545 | Acetamide | MDA-MB-231 | C71(0.74) | LDD2138 | [4] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C71(0.51) | LDD2113 | [4] |
| LDCM0156 | Aniline | NCI-H1299 | 13.19 | LDD0403 | [1] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C71(0.65); C144(0.79) | LDD2091 | [4] |
| LDCM0108 | Chloroacetamide | HeLa | C71(0.00); C144(0.00) | LDD0222 | [18] |
| LDCM0632 | CL-Sc | Hep-G2 | C144(20.00); C71(0.53) | LDD2227 | [20] |
| LDCM0625 | F8 | Ramos | C144(0.93) | LDD2187 | [21] |
| LDCM0572 | Fragment10 | Ramos | C144(0.53) | LDD2189 | [21] |
| LDCM0573 | Fragment11 | Ramos | C144(0.05) | LDD2190 | [21] |
| LDCM0574 | Fragment12 | Ramos | C144(0.55) | LDD2191 | [21] |
| LDCM0575 | Fragment13 | Ramos | C144(1.04) | LDD2192 | [21] |
| LDCM0576 | Fragment14 | Ramos | C144(0.85) | LDD2193 | [21] |
| LDCM0579 | Fragment20 | Ramos | C144(0.42) | LDD2194 | [21] |
| LDCM0580 | Fragment21 | Ramos | C144(0.95) | LDD2195 | [21] |
| LDCM0582 | Fragment23 | Ramos | C144(0.81) | LDD2196 | [21] |
| LDCM0578 | Fragment27 | Ramos | C144(1.22) | LDD2197 | [21] |
| LDCM0588 | Fragment30 | Ramos | C144(1.38) | LDD2199 | [21] |
| LDCM0589 | Fragment31 | Ramos | C144(1.44) | LDD2200 | [21] |
| LDCM0590 | Fragment32 | Ramos | C144(0.64) | LDD2201 | [21] |
| LDCM0468 | Fragment33 | Ramos | C144(1.31) | LDD2202 | [21] |
| LDCM0596 | Fragment38 | Ramos | C144(0.68) | LDD2203 | [21] |
| LDCM0566 | Fragment4 | Ramos | C144(0.63) | LDD2184 | [21] |
| LDCM0614 | Fragment56 | Ramos | C144(1.19) | LDD2205 | [21] |
| LDCM0569 | Fragment7 | Ramos | C144(0.49) | LDD2186 | [21] |
| LDCM0571 | Fragment9 | Ramos | C144(0.43) | LDD2188 | [21] |
| LDCM0116 | HHS-0101 | DM93 | Y115(0.92) | LDD0264 | [8] |
| LDCM0117 | HHS-0201 | DM93 | Y115(0.85) | LDD0265 | [8] |
| LDCM0118 | HHS-0301 | DM93 | Y115(0.89) | LDD0266 | [8] |
| LDCM0119 | HHS-0401 | DM93 | Y115(0.96) | LDD0267 | [8] |
| LDCM0120 | HHS-0701 | DM93 | Y115(0.95) | LDD0268 | [8] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [18] |
| LDCM0022 | KB02 | HEK-293T | C144(1.03) | LDD1492 | [22] |
| LDCM0023 | KB03 | HEK-293T | C144(0.94) | LDD1497 | [22] |
| LDCM0024 | KB05 | COLO792 | C94(2.77); C218(1.55) | LDD3310 | [7] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C71(1.47) | LDD2102 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [18] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C144(0.58) | LDD2089 | [4] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C71(1.12) | LDD2090 | [4] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C71(0.90) | LDD2093 | [4] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C71(2.42) | LDD2094 | [4] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C71(0.07) | LDD2096 | [4] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C71(0.61) | LDD2097 | [4] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C71(0.88) | LDD2098 | [4] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C71(1.01) | LDD2099 | [4] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C71(0.33) | LDD2100 | [4] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C71(0.83) | LDD2101 | [4] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C71(0.31) | LDD2104 | [4] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C71(0.16) | LDD2106 | [4] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C71(0.87) | LDD2107 | [4] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C71(0.36) | LDD2108 | [4] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C71(0.40) | LDD2109 | [4] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C71(0.88) | LDD2111 | [4] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C71(0.64) | LDD2114 | [4] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C71(0.33) | LDD2115 | [4] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C71(0.31) | LDD2116 | [4] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C71(0.24) | LDD2118 | [4] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C71(2.21) | LDD2119 | [4] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C71(0.88) | LDD2120 | [4] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C71(0.12) | LDD2122 | [4] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C71(0.12) | LDD2124 | [4] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C71(0.95) | LDD2125 | [4] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C71(0.09) | LDD2126 | [4] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C71(0.87) | LDD2127 | [4] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C71(1.01) | LDD2128 | [4] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C71(0.89) | LDD2129 | [4] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C71(0.32) | LDD2134 | [4] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C71(1.04) | LDD2135 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C71(0.90) | LDD2136 | [4] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C71(0.92) | LDD2137 | [4] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C71(2.11) | LDD1700 | [4] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C71(0.68) | LDD2140 | [4] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C71(0.67) | LDD2141 | [4] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C71(1.04) | LDD2143 | [4] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C71(3.07) | LDD2144 | [4] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C71(0.11) | LDD2145 | [4] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C71(0.72) | LDD2146 | [4] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C71(0.38) | LDD2148 | [4] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C71(0.11) | LDD2149 | [4] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C71(0.58) | LDD2150 | [4] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C71(0.11) | LDD2151 | [4] |
| LDCM0131 | RA190 | MM1.R | C144(1.04) | LDD0304 | [23] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| X-box-binding protein 1 (XBP1) | BZIP family | P17861 | |||
Other
References
