Details of the Target
General Information of Target
| Target ID | LDTP05458 | |||||
|---|---|---|---|---|---|---|
| Target Name | Serine/arginine-rich splicing factor 11 (SRSF11) | |||||
| Gene Name | SRSF11 | |||||
| Gene ID | 9295 | |||||
| Synonyms |
SFRS11; Serine/arginine-rich splicing factor 11; Arginine-rich 54 kDa nuclear protein; p54; Splicing factor, arginine/serine-rich 11 |
|||||
| 3D Structure | ||||||
| Sequence |
MSNTTVVPSTAGPGPSGGPGGGGGGGGGGGGTEVIQVTNVSPSASSEQMRTLFGFLGKID
ELRLFPPDDSPLPVSSRVCFVKFHDPDSAVVAQHLTNTVFVDRALIVVPYAEGVIPDEAK ALSLLAPANAVAGLLPGGGLLPTPNPLTQIGAVPLAALGAPTLDPALAALGLPGANLNSQ SLAADQLLKLMSTVDPKLNHVAAGLVSPSLKSDTSSKEIEEAMKRVREAQSLISAAIEPD KKEEKRRHSRSRSRSRRRRTPSSSRHRRSRSRSRRRSHSKSRSRRRSKSPRRRRSHSRER GRRSRSTSKTRDKKKEDKEKKRSKTPPKSYSTARRSRSASRERRRRRSRSGTRSPKKPRS PKRKLSRSPSPRRHKKEKKKDKDKERSRDERERSTSKKKKSKDKEKDRERKSESDKDVKQ VTRDYDEEEQGYDSEKEKKEEKKPIETGSPKTKECSVEKGTGDSLRESKVNGDDHHEEDM DMSD |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
Splicing factor SR family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function | May function in pre-mRNA splicing. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| AN3CA | SNV: p.S9R; p.R254C | . | |||
| ETK1 | SNV: p.E47Q | DBIA Probe Info | |||
| HT115 | SNV: p.R393Ter | . | |||
| HUH28 | SNV: p.E377K | . | |||
| OVCAR4 | SNV: p.E478K | . | |||
| SNGM | SNV: p.R372G | . | |||
| SSP25 | SNV: p.E47Q | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
8.70 | LDD0402 | [1] | |
|
C-Sul Probe Info |
 |
6.54 | LDD0066 | [2] | |
|
STPyne Probe Info |
 |
K197(1.06); K211(3.61); K459(6.67) | LDD0277 | [3] | |
|
Probe 1 Probe Info |
 |
Y425(122.37) | LDD3495 | [4] | |
|
DBIA Probe Info |
 |
C455(1.25) | LDD3312 | [5] | |
|
BTD Probe Info |
 |
C455(0.54) | LDD2089 | [6] | |
|
5E-2FA Probe Info |
 |
H94(0.00); H200(0.00) | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
K459(0.00); K211(0.00); K217(0.00); K453(0.00) | LDD0199 | [8] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [9] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [9] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [10] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [11] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [10] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [12] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [13] | |
|
Acrolein Probe Info |
 |
H84(0.00); H200(0.00); H94(0.00) | LDD0217 | [14] | |
|
TER-AC Probe Info |
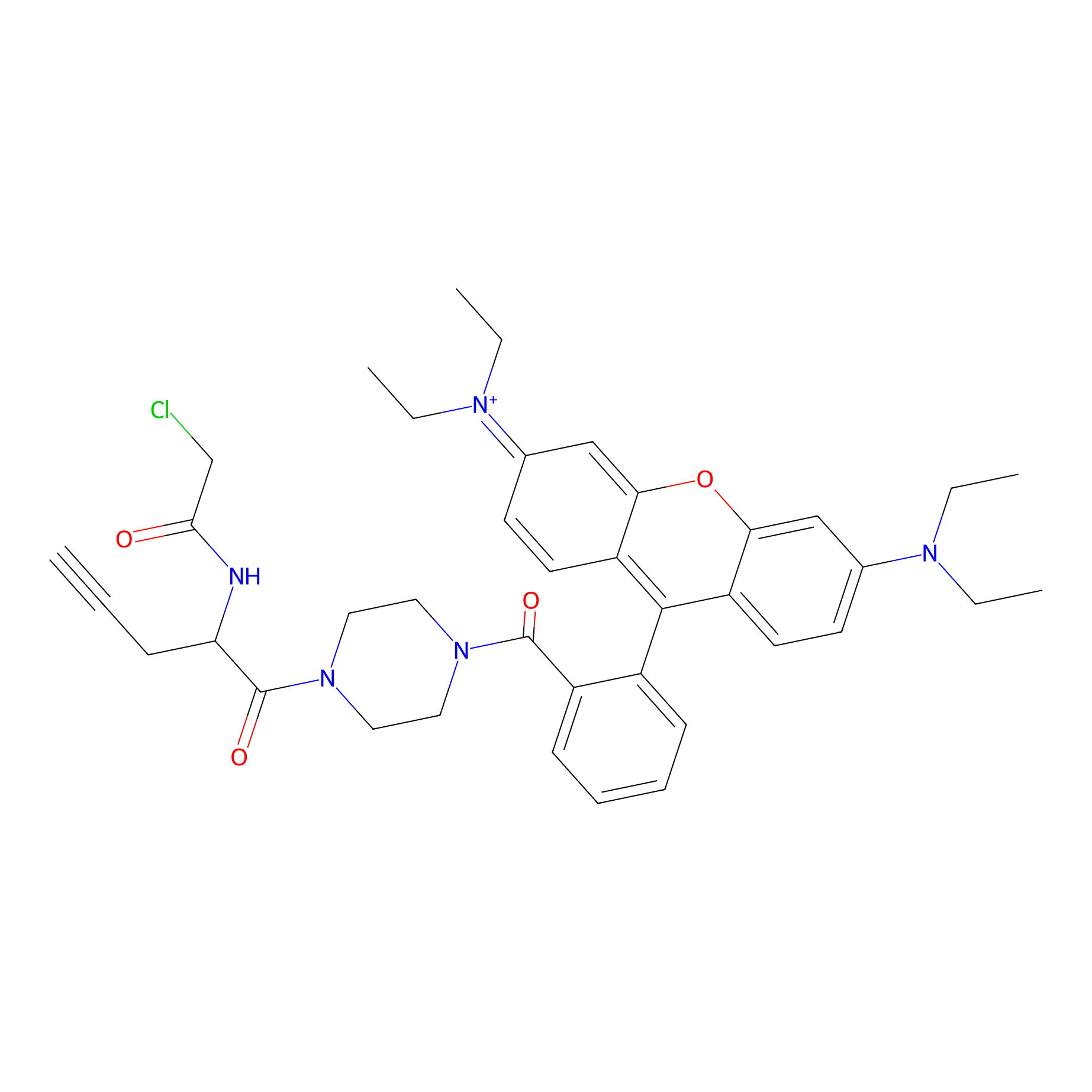 |
N.A. | LDD0426 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C455(0.73) | LDD2142 | [6] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C455(0.89) | LDD2112 | [6] |
| LDCM0502 | 1-(Cyanoacetyl)piperidine | MDA-MB-231 | C455(0.95) | LDD2095 | [6] |
| LDCM0537 | 2-Cyano-N,N-dimethylacetamide | MDA-MB-231 | C455(0.97) | LDD2130 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C455(0.73) | LDD2117 | [6] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C455(1.07) | LDD2152 | [6] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C455(0.83) | LDD2103 | [6] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C455(0.73) | LDD2132 | [6] |
| LDCM0538 | 4-(Cyanoacetyl)morpholine | MDA-MB-231 | C455(0.71) | LDD2131 | [6] |
| LDCM0545 | Acetamide | MDA-MB-231 | C455(0.42) | LDD2138 | [6] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C455(0.58) | LDD2091 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | H84(0.00); H200(0.00) | LDD0222 | [14] |
| LDCM0625 | F8 | Ramos | C455(1.57) | LDD2187 | [16] |
| LDCM0572 | Fragment10 | Ramos | C455(0.65) | LDD2189 | [16] |
| LDCM0573 | Fragment11 | Ramos | C455(1.31) | LDD2190 | [16] |
| LDCM0574 | Fragment12 | Ramos | C455(0.55) | LDD2191 | [16] |
| LDCM0575 | Fragment13 | Ramos | C455(0.74) | LDD2192 | [16] |
| LDCM0576 | Fragment14 | Ramos | C455(0.56) | LDD2193 | [16] |
| LDCM0579 | Fragment20 | Ramos | C455(0.49) | LDD2194 | [16] |
| LDCM0580 | Fragment21 | Ramos | C455(0.68) | LDD2195 | [16] |
| LDCM0582 | Fragment23 | Ramos | C455(0.54) | LDD2196 | [16] |
| LDCM0578 | Fragment27 | Ramos | C455(0.95) | LDD2197 | [16] |
| LDCM0586 | Fragment28 | Ramos | C455(0.84) | LDD2198 | [16] |
| LDCM0588 | Fragment30 | Ramos | C455(1.37) | LDD2199 | [16] |
| LDCM0589 | Fragment31 | Ramos | C455(0.69) | LDD2200 | [16] |
| LDCM0590 | Fragment32 | Ramos | C455(0.93) | LDD2201 | [16] |
| LDCM0468 | Fragment33 | Ramos | C455(1.25) | LDD2202 | [16] |
| LDCM0596 | Fragment38 | Ramos | C455(0.80) | LDD2203 | [16] |
| LDCM0566 | Fragment4 | Ramos | C455(0.80) | LDD2184 | [16] |
| LDCM0610 | Fragment52 | Ramos | C455(1.07) | LDD2204 | [16] |
| LDCM0614 | Fragment56 | Ramos | C455(1.23) | LDD2205 | [16] |
| LDCM0569 | Fragment7 | Ramos | C455(0.78) | LDD2186 | [16] |
| LDCM0571 | Fragment9 | Ramos | C455(0.48) | LDD2188 | [16] |
| LDCM0107 | IAA | HeLa | H84(0.00); H200(0.00) | LDD0221 | [14] |
| LDCM0022 | KB02 | Ramos | C455(0.92) | LDD2182 | [16] |
| LDCM0023 | KB03 | Ramos | C455(0.66) | LDD2183 | [16] |
| LDCM0024 | KB05 | HMCB | C455(1.25) | LDD3312 | [5] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C455(0.84) | LDD2102 | [6] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C455(0.39) | LDD2121 | [6] |
| LDCM0109 | NEM | HeLa | H84(0.00); H200(0.00); H94(0.00) | LDD0223 | [14] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C455(0.54) | LDD2089 | [6] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C455(1.02) | LDD2090 | [6] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C455(0.66) | LDD2093 | [6] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C455(1.30) | LDD2094 | [6] |
| LDCM0503 | Nucleophilic fragment 14b | MDA-MB-231 | C455(1.23) | LDD2096 | [6] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C455(0.67) | LDD2099 | [6] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C455(0.96) | LDD2100 | [6] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C455(1.01) | LDD2104 | [6] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C455(1.27) | LDD2105 | [6] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C455(0.87) | LDD2107 | [6] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C455(0.76) | LDD2108 | [6] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C455(0.75) | LDD2109 | [6] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C455(1.68) | LDD2110 | [6] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C455(0.66) | LDD2111 | [6] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C455(0.89) | LDD2114 | [6] |
| LDCM0523 | Nucleophilic fragment 24b | MDA-MB-231 | C455(1.18) | LDD2116 | [6] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C455(1.21) | LDD2118 | [6] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C455(2.27) | LDD2119 | [6] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C455(1.10) | LDD2120 | [6] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C455(1.26) | LDD2122 | [6] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C455(0.67) | LDD2123 | [6] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C455(1.27) | LDD2124 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C455(0.55) | LDD2125 | [6] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C455(1.17) | LDD2126 | [6] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C455(0.76) | LDD2127 | [6] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C455(0.94) | LDD2128 | [6] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C455(0.54) | LDD2129 | [6] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C455(0.44) | LDD2135 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C455(0.83) | LDD2136 | [6] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C455(0.90) | LDD2137 | [6] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C455(0.64) | LDD2140 | [6] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C455(0.71) | LDD2141 | [6] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C455(1.47) | LDD2143 | [6] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C455(3.83) | LDD2144 | [6] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C455(3.50) | LDD2145 | [6] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C455(0.68) | LDD2146 | [6] |
| LDCM0553 | Nucleophilic fragment 6b | MDA-MB-231 | C455(6.76) | LDD2147 | [6] |
| LDCM0555 | Nucleophilic fragment 7b | MDA-MB-231 | C455(1.59) | LDD2149 | [6] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C455(0.32) | LDD2150 | [6] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C455(1.77) | LDD2151 | [6] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C455(1.14) | LDD2153 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| ATP synthase subunit delta, mitochondrial (ATP5F1D) | ATPase epsilon chain family | P30049 | |||
| Syntenin-1 (SDCBP) | . | O00560 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protein C-ets-1 (ETS1) | ETS family | P14921 | |||
| Paired box protein Pax-6 (PAX6) | Paired homeobox family | P26367 | |||
Other
References
