Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Alkylaryl probe 2 Probe Info |
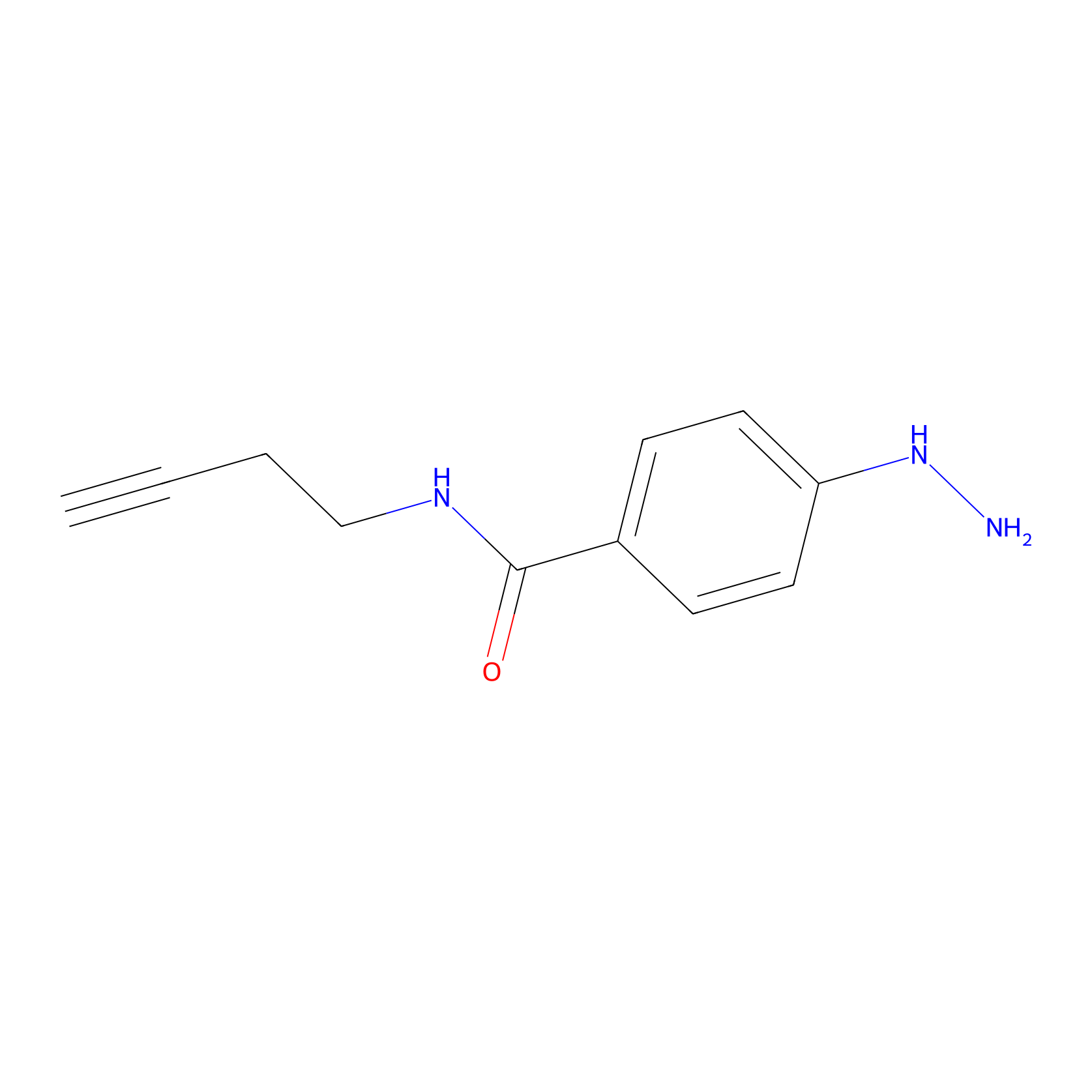 |
20.00 | LDD0391 | [1] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [2] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [2] | |
|
DBIA Probe Info |
 |
C427(2.04) | LDD3410 | [3] | |
|
Jackson_14 Probe Info |
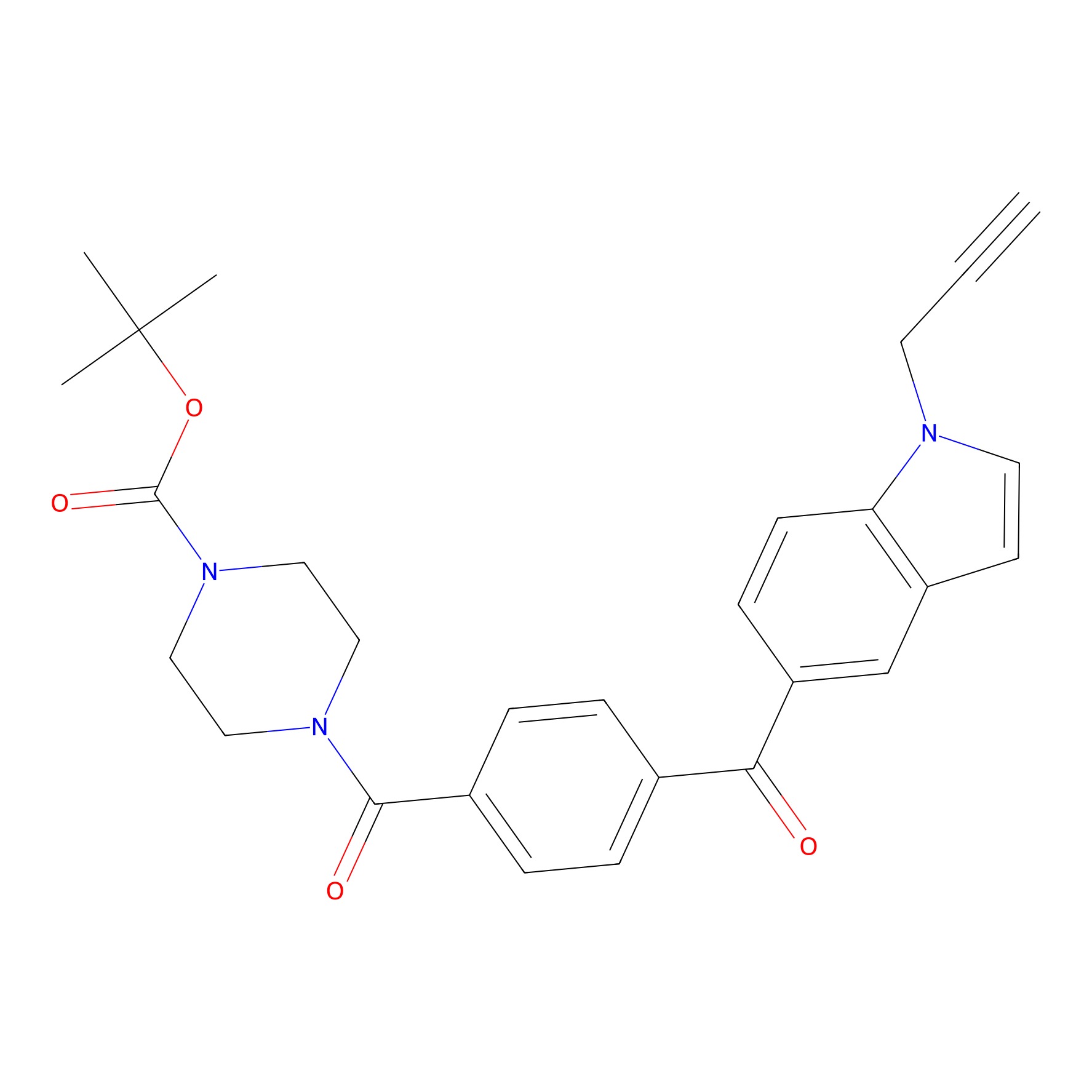 |
2.27 | LDD0123 | [4] | |
|
CY-1 Probe Info |
 |
N.A. | LDD0246 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [6] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
PARPYnD Probe Info |
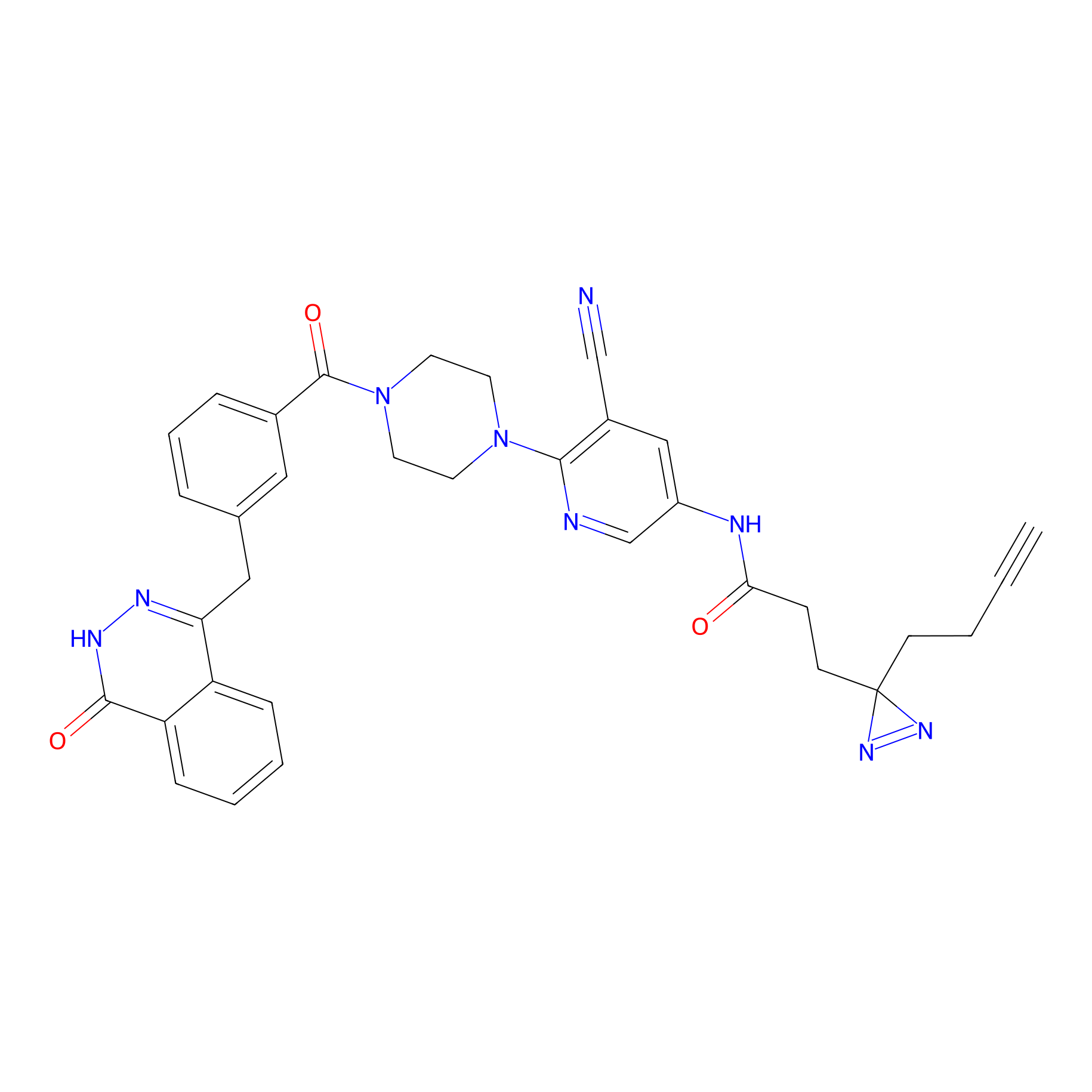 |
1.74 | LDD0374 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0147 | AZ0108 | MDA-MB-468 | 1.46 | LDD0376 | [8] |
| LDCM0022 | KB02 | A101D | C427(1.61) | LDD2250 | [3] |
| LDCM0023 | KB03 | A101D | C427(3.48) | LDD2667 | [3] |
| LDCM0024 | KB05 | RPMI-7951 | C427(2.04) | LDD3410 | [3] |
| LDCM0099 | Phenelzine | MDA-MB-231 | 3.00 | LDD0392 | [1] |
| LDCM0016 | Ranjitkar_cp1 | MDA-MB-231 | 2.27 | LDD0123 | [4] |
The Interaction Atlas With This Target
References
