Details of the Target
General Information of Target
| Target ID | LDTP04992 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ras-related protein Rab-11A (RAB11A) | |||||
| Gene Name | RAB11A | |||||
| Gene ID | 8766 | |||||
| Synonyms |
RAB11; Ras-related protein Rab-11A; Rab-11; EC 3.6.5.2; YL8 |
|||||
| 3D Structure | ||||||
| Sequence |
MGTRDDEYDYLFKVVLIGDSGVGKSNLLSRFTRNEFNLESKSTIGVEFATRSIQVDGKTI
KAQIWDTAGQERYRAITSAYYRGAVGALLVYDIAKHLTYENVERWLKELRDHADSNIVIM LVGNKSDLRHLRAVPTDEARAFAEKNGLSFIETSALDSTNVEAAFQTILTEIYRIVSQKQ MSDRRENDMSPSNNVVPIHVPPTTENKPKVQCCQNI |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Small GTPase superfamily, Rab family
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function |
The small GTPases Rab are key regulators of intracellular membrane trafficking, from the formation of transport vesicles to their fusion with membranes. Rabs cycle between an inactive GDP-bound form and an active GTP-bound form that is able to recruit to membranes different set of downstream effectors directly responsible for vesicle formation, movement, tethering and fusion. The small Rab GTPase RAB11A regulates endocytic recycling. Forms a functional Rab11/FIP3/dynein complex that regulates the movement of peripheral sorting endosomes (SE) along microtubule tracks toward the microtubule organizing center/centrosome, generating the endosomal recycling compartment (ERC). Acts as a major regulator of membrane delivery during cytokinesis. Together with MYO5B and RAB8A participates in epithelial cell polarization. Together with RAB3IP, RAB8A, the exocyst complex, PARD3, PRKCI, ANXA2, CDC42 and DNMBP promotes transcytosis of PODXL to the apical membrane initiation sites (AMIS), apical surface formation and lumenogenesis. Together with MYO5B participates in CFTR trafficking to the plasma membrane and TF (Transferrin) recycling in nonpolarized cells. Required in a complex with MYO5B and RAB11FIP2 for the transport of NPC1L1 to the plasma membrane. Participates in the sorting and basolateral transport of CDH1 from the Golgi apparatus to the plasma membrane. Regulates the recycling of FCGRT (receptor of Fc region of monomeric Ig G) to basolateral membranes. May also play a role in melanosome transport and release from melanocytes. Promotes Rabin8/RAB3IP preciliary vesicular trafficking to mother centriole by forming a ciliary targeting complex containing Rab11, ASAP1, Rabin8/RAB3IP, RAB11FIP3 and ARF4, thereby regulating ciliogenesis initiation. On the contrary, upon LPAR1 receptor signaling pathway activation, interaction with phosphorylated WDR44 prevents Rab11-RAB3IP-RAB11FIP3 complex formation and cilia growth. Participates in the export of a subset of neosynthesized proteins through a Rab8-Rab10-Rab11-endososomal dependent export route via interaction with WDR44.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
AZ-9 Probe Info |
 |
10.00 | LDD0393 | [1] | |
|
C-Sul Probe Info |
 |
4.54 | LDD0066 | [2] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [3] | |
|
STPyne Probe Info |
 |
K179(5.16) | LDD0277 | [4] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [4] | |
|
OPA-S-S-alkyne Probe Info |
 |
K179(2.31); K61(3.24) | LDD3494 | [5] | |
|
Probe 1 Probe Info |
 |
Y8(21.21) | LDD3495 | [6] | |
|
HPAP Probe Info |
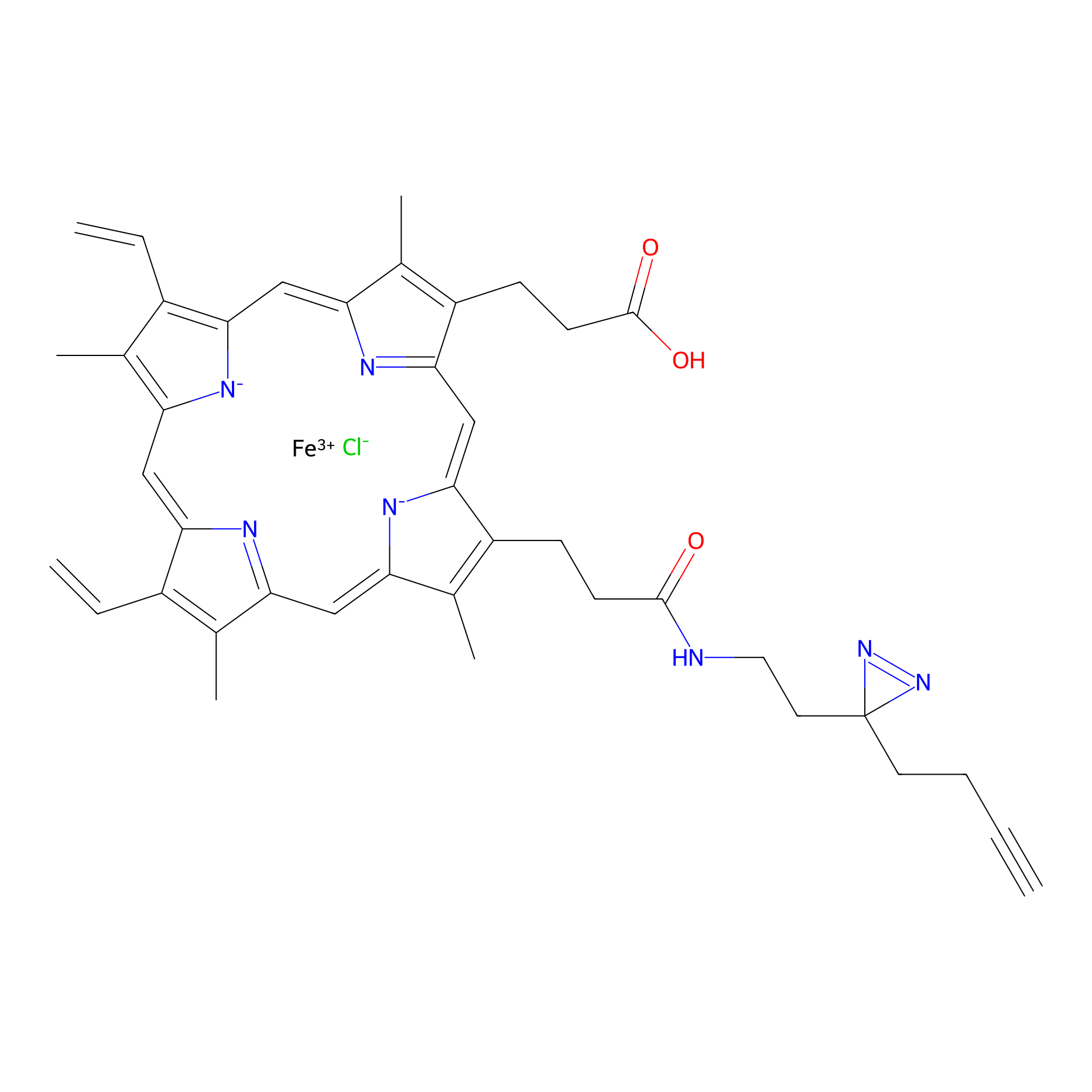 |
3.70 | LDD0062 | [7] | |
|
5E-2FA Probe Info |
 |
H112(0.00); H199(0.00) | LDD2235 | [8] | |
|
ATP probe Probe Info |
 |
K58(0.00); K61(0.00); K179(0.00) | LDD0199 | [9] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [8] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [10] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [11] | |
|
SF Probe Info |
 |
Y8(0.00); Y10(0.00) | LDD0028 | [12] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DR-1 Probe Info |
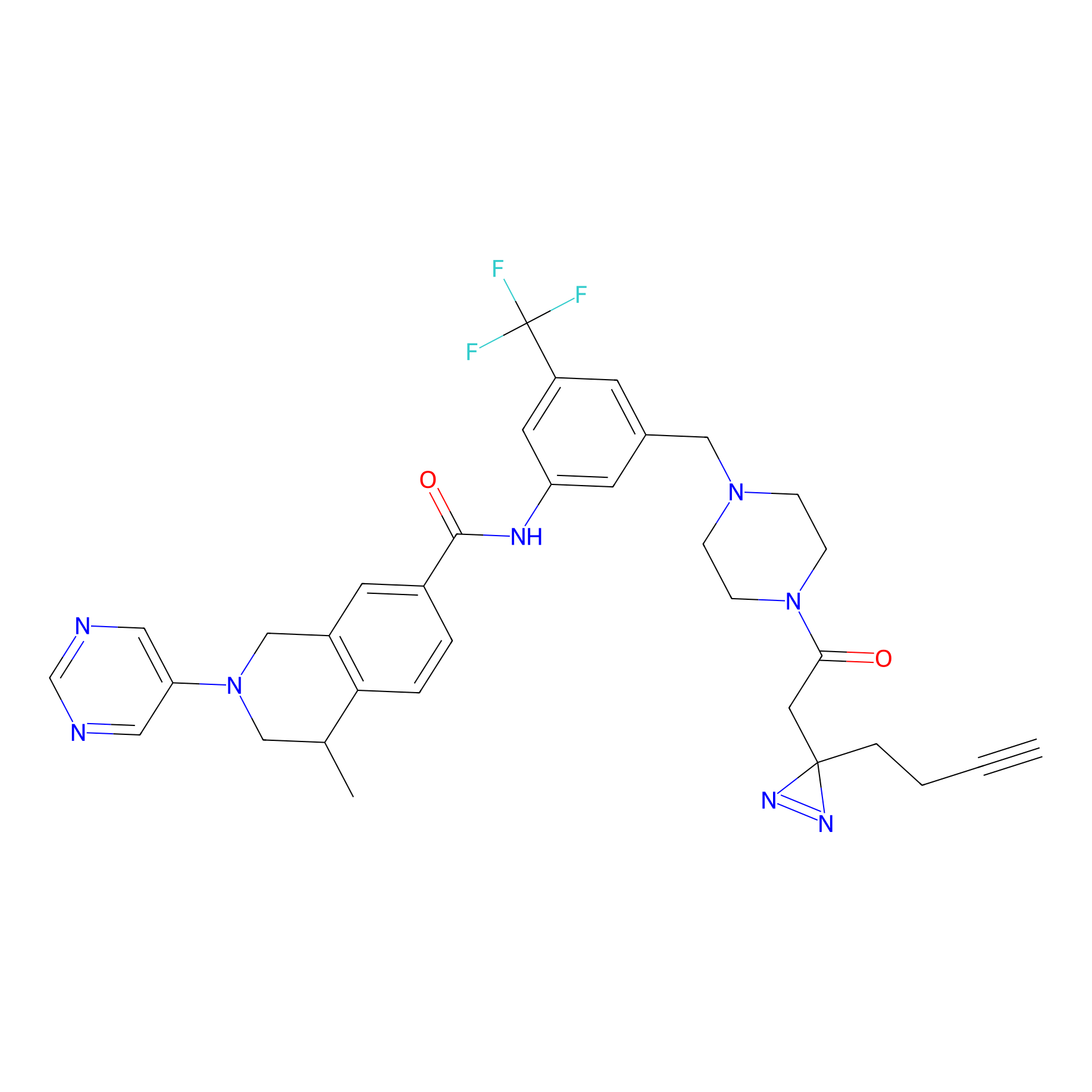 |
5.29 | LDD0398 | [15] | |
|
FFF probe11 Probe Info |
 |
20.00 | LDD0471 | [16] | |
|
FFF probe13 Probe Info |
 |
20.00 | LDD0475 | [16] | |
|
FFF probe14 Probe Info |
 |
19.06 | LDD0477 | [16] | |
|
FFF probe2 Probe Info |
 |
20.00 | LDD0463 | [16] | |
|
FFF probe3 Probe Info |
 |
20.00 | LDD0464 | [16] | |
|
FFF probe4 Probe Info |
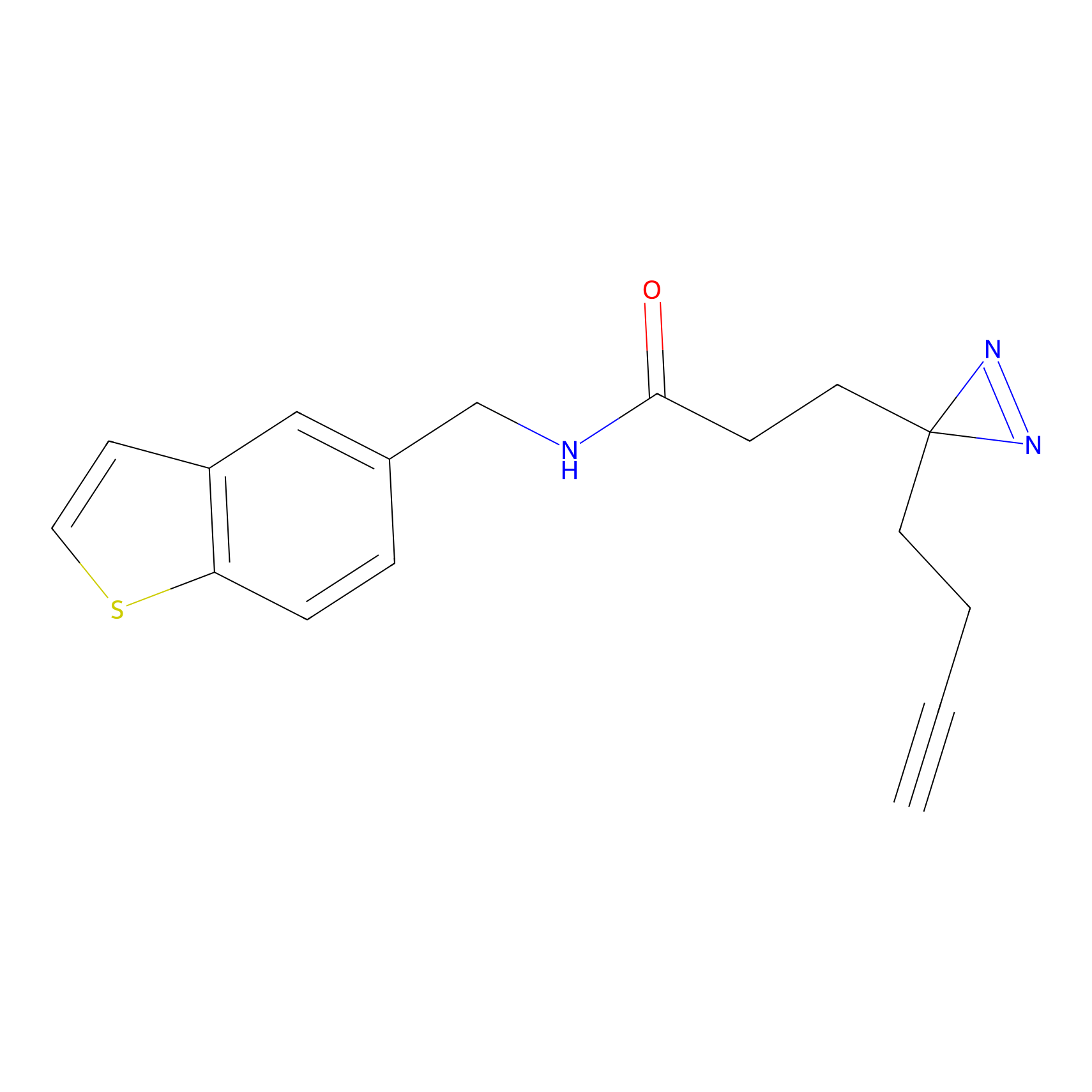 |
20.00 | LDD0466 | [16] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Protrudin (ZFYVE27) | . | Q5T4F4 | |||
Transcription factor
Other
References
