Details of the Target
General Information of Target
| Target ID | LDTP03828 | |||||
|---|---|---|---|---|---|---|
| Target Name | Sterol regulatory element-binding protein 1 (SREBF1) | |||||
| Gene Name | SREBF1 | |||||
| Gene ID | 6720 | |||||
| Synonyms |
BHLHD1; SREBP1; Sterol regulatory element-binding protein 1; SREBP-1; Class D basic helix-loop-helix protein 1; bHLHd1; Sterol regulatory element-binding transcription factor 1) [Cleaved into: Processed sterol regulatory element-binding protein 1; Transcription factor SREBF1)]
|
|||||
| 3D Structure | ||||||
| Sequence |
MDEPPFSEAALEQALGEPCDLDAALLTDIEDMLQLINNQDSDFPGLFDPPYAGSGAGGTD
PASPDTSSPGSLSPPPATLSSSLEAFLSGPQAAPSPLSPPQPAPTPLKMYPSMPAFSPGP GIKEESVPLSILQTPTPQPLPGALLPQSFPAPAPPQFSSTPVLGYPSPPGGFSTGSPPGN TQQPLPGLPLASPPGVPPVSLHTQVQSVVPQQLLTVTAAPTAAPVTTTVTSQIQQVPVLL QPHFIKADSLLLTAMKTDGATVKAAGLSPLVSGTTVQTGPLPTLVSGGTILATVPLVVDA EKLPINRLAAGSKAPASAQSRGEKRTAHNAIEKRYRSSINDKIIELKDLVVGTEAKLNKS AVLRKAIDYIRFLQHSNQKLKQENLSLRTAVHKSKSLKDLVSACGSGGNTDVLMEGVKTE VEDTLTPPPSDAGSPFQSSPLSLGSRGSGSGGSGSDSEPDSPVFEDSKAKPEQRPSLHSR GMLDRSRLALCTLVFLCLSCNPLASLLGARGLPSPSDTTSVYHSPGRNVLGTESRDGPGW AQWLLPPVVWLLNGLLVLVSLVLLFVYGEPVTRPHSGPAVYFWRHRKQADLDLARGDFAQ AAQQLWLALRALGRPLPTSHLDLACSLLWNLIRHLLQRLWVGRWLAGRAGGLQQDCALRV DASASARDAALVYHKLHQLHTMGKHTGGHLTATNLALSALNLAECAGDAVSVATLAEIYV AAALRVKTSLPRALHFLTRFFLSSARQACLAQSGSVPPAMQWLCHPVGHRFFVDGDWSVL STPWESLYSLAGNPVDPLAQVTQLFREHLLERALNCVTQPNPSPGSADGDKEFSDALGYL QLLNSCSDAAGAPAYSFSISSSMATTTGVDPVAKWWASLTAVVIHWLRRDEEAAERLCPL VEHLPRVLQESERPLPRAALHSFKAARALLGCAKAESGPASLTICEKASGYLQDSLATTP ASSSIDKAVQLFLCDLLLVVRTSLWRQQQPPAPAPAAQGTSSRPQASALELRGFQRDLSS LRRLAQSFRPAMRRVFLHEATARLMAGASPTRTHQLLDRSLRRRAGPGGKGGAVAELEPR PTRREHAEALLLASCYLPPGFLSAPGQRVGMLAEAARTLEKLGDRRLLHDCQQMLMRLGG GTTVTSS |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Transcription factor
|
|||||
| Family |
SREBP family
|
|||||
| Subcellular location |
Nucleus; Endoplasmic reticulum membrane
|
|||||
| Function |
[Sterol regulatory element-binding protein 1]: Precursor of the transcription factor form (Processed sterol regulatory element-binding protein 1), which is embedded in the endoplasmic reticulum membrane. Low sterol concentrations promote processing of this form, releasing the transcription factor form that translocates into the nucleus and activates transcription of genes involved in cholesterol biosynthesis and lipid homeostasis.; [Processed sterol regulatory element-binding protein 1]: Key transcription factor that regulates expression of genes involved in cholesterol biosynthesis and lipid homeostasis. Binds to the sterol regulatory element 1 (SRE-1) (5'-ATCACCCCAC-3'). Has dual sequence specificity binding to both an E-box motif (5'-ATCACGTGA-3') and to SRE-1 (5'-ATCACCCCAC-3'). Regulates the promoters of genes involved in cholesterol biosynthesis and the LDL receptor (LDLR) pathway of sterol regulation.; [Isoform SREBP-1A]: Isoform expressed only in select tissues, which has higher transcriptional activity compared to SREBP-1C. Able to stimulate both lipogenic and cholesterogenic gene expression. Has a role in the nutritional regulation of fatty acids and triglycerides in lipogenic organs such as the liver. Required for innate immune response in macrophages by regulating lipid metabolism.; [Isoform SREBP-1C]: Predominant isoform expressed in most tissues, which has weaker transcriptional activity compared to isoform SREBP-1A. Primarily controls expression of lipogenic gene. Strongly activates global lipid synthesis in rapidly growing cells.; [Isoform SREBP-1aDelta]: The absence of Golgi proteolytic processing requirement makes this isoform constitutively active in transactivation of lipogenic gene promoters.; [Isoform SREBP-1cDelta]: The absence of Golgi proteolytic processing requirement makes this isoform constitutively active in transactivation of lipogenic gene promoters.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
10.42 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K365(10.00) | LDD0277 | [2] | |
|
DBIA Probe Info |
 |
C962(15.26) | LDD3311 | [3] | |
|
NAIA_4 Probe Info |
 |
C945(0.00); C1095(0.00) | LDD2226 | [4] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2224 | [4] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [5] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [6] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [6] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [5] | |
|
TFBX Probe Info |
 |
C656(0.00); C932(0.00); C898(0.00) | LDD0148 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Alk-rapa Probe Info |
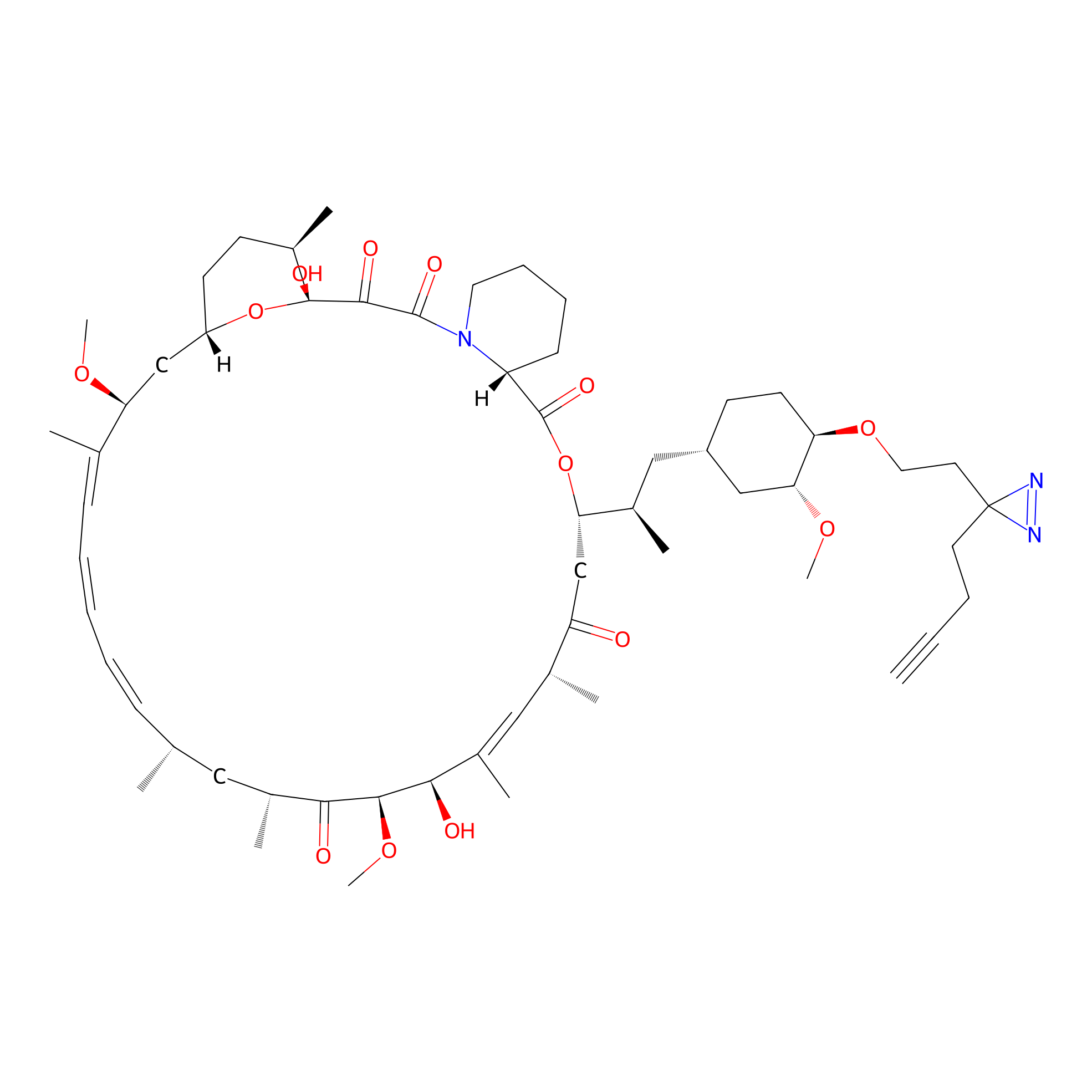 |
8.80 | LDD0213 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C932(1.08) | LDD1507 | [9] |
| LDCM0215 | AC10 | HEK-293T | C932(0.93) | LDD1508 | [9] |
| LDCM0226 | AC11 | HEK-293T | C932(0.94) | LDD1509 | [9] |
| LDCM0237 | AC12 | HEK-293T | C932(1.16) | LDD1510 | [9] |
| LDCM0259 | AC14 | HEK-293T | C932(0.86) | LDD1512 | [9] |
| LDCM0270 | AC15 | HEK-293T | C932(1.01); C816(1.34) | LDD1513 | [9] |
| LDCM0276 | AC17 | HEK-293T | C932(1.05) | LDD1515 | [9] |
| LDCM0277 | AC18 | HEK-293T | C932(1.02) | LDD1516 | [9] |
| LDCM0278 | AC19 | HEK-293T | C932(1.00) | LDD1517 | [9] |
| LDCM0279 | AC2 | HEK-293T | C932(0.93) | LDD1518 | [9] |
| LDCM0280 | AC20 | HEK-293T | C932(1.06) | LDD1519 | [9] |
| LDCM0282 | AC22 | HEK-293T | C932(0.97) | LDD1521 | [9] |
| LDCM0283 | AC23 | HEK-293T | C932(1.00); C816(1.21) | LDD1522 | [9] |
| LDCM0284 | AC24 | HEK-293T | C932(0.92) | LDD1523 | [9] |
| LDCM0285 | AC25 | HEK-293T | C932(1.09) | LDD1524 | [9] |
| LDCM0286 | AC26 | HEK-293T | C932(0.93) | LDD1525 | [9] |
| LDCM0287 | AC27 | HEK-293T | C932(0.87) | LDD1526 | [9] |
| LDCM0288 | AC28 | HEK-293T | C932(0.91) | LDD1527 | [9] |
| LDCM0290 | AC3 | HEK-293T | C932(1.07) | LDD1529 | [9] |
| LDCM0291 | AC30 | HEK-293T | C932(0.89) | LDD1530 | [9] |
| LDCM0292 | AC31 | HEK-293T | C932(1.02); C816(1.17) | LDD1531 | [9] |
| LDCM0293 | AC32 | HEK-293T | C932(0.96) | LDD1532 | [9] |
| LDCM0294 | AC33 | HEK-293T | C932(1.23) | LDD1533 | [9] |
| LDCM0295 | AC34 | HEK-293T | C932(1.02) | LDD1534 | [9] |
| LDCM0296 | AC35 | HEK-293T | C932(0.96) | LDD1535 | [9] |
| LDCM0297 | AC36 | HEK-293T | C932(1.03) | LDD1536 | [9] |
| LDCM0299 | AC38 | HEK-293T | C932(0.98) | LDD1538 | [9] |
| LDCM0300 | AC39 | HEK-293T | C932(1.04); C816(1.20) | LDD1539 | [9] |
| LDCM0301 | AC4 | HEK-293T | C932(1.02) | LDD1540 | [9] |
| LDCM0302 | AC40 | HEK-293T | C932(0.88) | LDD1541 | [9] |
| LDCM0303 | AC41 | HEK-293T | C932(1.17) | LDD1542 | [9] |
| LDCM0304 | AC42 | HEK-293T | C932(0.98) | LDD1543 | [9] |
| LDCM0305 | AC43 | HEK-293T | C932(0.98) | LDD1544 | [9] |
| LDCM0306 | AC44 | HEK-293T | C932(1.05) | LDD1545 | [9] |
| LDCM0308 | AC46 | HEK-293T | C932(0.87) | LDD1547 | [9] |
| LDCM0309 | AC47 | HEK-293T | C932(1.05); C816(1.01) | LDD1548 | [9] |
| LDCM0310 | AC48 | HEK-293T | C932(0.75) | LDD1549 | [9] |
| LDCM0311 | AC49 | HEK-293T | C932(1.06) | LDD1550 | [9] |
| LDCM0313 | AC50 | HEK-293T | C932(0.92) | LDD1552 | [9] |
| LDCM0314 | AC51 | HEK-293T | C932(0.88) | LDD1553 | [9] |
| LDCM0315 | AC52 | HEK-293T | C932(1.09) | LDD1554 | [9] |
| LDCM0317 | AC54 | HEK-293T | C932(0.80) | LDD1556 | [9] |
| LDCM0318 | AC55 | HEK-293T | C932(1.00); C816(1.26) | LDD1557 | [9] |
| LDCM0319 | AC56 | HEK-293T | C932(0.80) | LDD1558 | [9] |
| LDCM0320 | AC57 | HEK-293T | C932(1.08) | LDD1559 | [9] |
| LDCM0321 | AC58 | HEK-293T | C932(0.84) | LDD1560 | [9] |
| LDCM0322 | AC59 | HEK-293T | C932(1.25) | LDD1561 | [9] |
| LDCM0323 | AC6 | HEK-293T | C932(0.91) | LDD1562 | [9] |
| LDCM0324 | AC60 | HEK-293T | C932(1.13) | LDD1563 | [9] |
| LDCM0326 | AC62 | HEK-293T | C932(0.86) | LDD1565 | [9] |
| LDCM0327 | AC63 | HEK-293T | C932(1.02); C816(1.18) | LDD1566 | [9] |
| LDCM0328 | AC64 | HEK-293T | C932(0.77) | LDD1567 | [9] |
| LDCM0334 | AC7 | HEK-293T | C932(1.00); C816(1.47) | LDD1568 | [9] |
| LDCM0345 | AC8 | HEK-293T | C932(0.96) | LDD1569 | [9] |
| LDCM0356 | AKOS034007680 | HEK-293T | C932(1.10) | LDD1570 | [9] |
| LDCM0275 | AKOS034007705 | HEK-293T | C932(0.85) | LDD1514 | [9] |
| LDCM0367 | CL1 | HEK-293T | C932(1.10); C404(1.10) | LDD1571 | [9] |
| LDCM0368 | CL10 | HEK-293T | C932(1.66) | LDD1572 | [9] |
| LDCM0369 | CL100 | HEK-293T | C932(0.87) | LDD1573 | [9] |
| LDCM0370 | CL101 | HEK-293T | C932(1.06); C404(1.13) | LDD1574 | [9] |
| LDCM0371 | CL102 | HEK-293T | C932(1.24) | LDD1575 | [9] |
| LDCM0372 | CL103 | HEK-293T | C932(1.06) | LDD1576 | [9] |
| LDCM0373 | CL104 | HEK-293T | C932(0.90) | LDD1577 | [9] |
| LDCM0374 | CL105 | HEK-293T | C932(1.06); C404(1.07) | LDD1578 | [9] |
| LDCM0375 | CL106 | HEK-293T | C932(1.40) | LDD1579 | [9] |
| LDCM0376 | CL107 | HEK-293T | C932(1.14) | LDD1580 | [9] |
| LDCM0377 | CL108 | HEK-293T | C932(1.14) | LDD1581 | [9] |
| LDCM0378 | CL109 | HEK-293T | C932(0.97); C404(1.09) | LDD1582 | [9] |
| LDCM0379 | CL11 | HEK-293T | C932(1.14); C816(1.02) | LDD1583 | [9] |
| LDCM0380 | CL110 | HEK-293T | C932(1.32) | LDD1584 | [9] |
| LDCM0381 | CL111 | HEK-293T | C932(0.96) | LDD1585 | [9] |
| LDCM0382 | CL112 | HEK-293T | C932(0.93) | LDD1586 | [9] |
| LDCM0383 | CL113 | HEK-293T | C932(1.04); C404(1.23) | LDD1587 | [9] |
| LDCM0384 | CL114 | HEK-293T | C932(1.44) | LDD1588 | [9] |
| LDCM0385 | CL115 | HEK-293T | C932(1.10) | LDD1589 | [9] |
| LDCM0386 | CL116 | HEK-293T | C932(0.94) | LDD1590 | [9] |
| LDCM0387 | CL117 | HEK-293T | C932(1.12); C404(1.14) | LDD1591 | [9] |
| LDCM0388 | CL118 | HEK-293T | C932(1.36) | LDD1592 | [9] |
| LDCM0389 | CL119 | HEK-293T | C932(1.03) | LDD1593 | [9] |
| LDCM0390 | CL12 | HEK-293T | C932(0.94) | LDD1594 | [9] |
| LDCM0391 | CL120 | HEK-293T | C932(0.95) | LDD1595 | [9] |
| LDCM0392 | CL121 | HEK-293T | C932(1.06); C404(1.15) | LDD1596 | [9] |
| LDCM0393 | CL122 | HEK-293T | C932(1.01) | LDD1597 | [9] |
| LDCM0394 | CL123 | HEK-293T | C932(1.20) | LDD1598 | [9] |
| LDCM0395 | CL124 | HEK-293T | C932(0.78) | LDD1599 | [9] |
| LDCM0396 | CL125 | HEK-293T | C932(1.14); C404(1.20) | LDD1600 | [9] |
| LDCM0397 | CL126 | HEK-293T | C932(1.21) | LDD1601 | [9] |
| LDCM0398 | CL127 | HEK-293T | C932(0.98) | LDD1602 | [9] |
| LDCM0399 | CL128 | HEK-293T | C932(0.93) | LDD1603 | [9] |
| LDCM0400 | CL13 | HEK-293T | C932(1.15); C404(1.13) | LDD1604 | [9] |
| LDCM0401 | CL14 | HEK-293T | C932(1.07) | LDD1605 | [9] |
| LDCM0402 | CL15 | HEK-293T | C932(1.30) | LDD1606 | [9] |
| LDCM0403 | CL16 | HEK-293T | C932(1.02) | LDD1607 | [9] |
| LDCM0404 | CL17 | HEK-293T | C932(1.33) | LDD1608 | [9] |
| LDCM0405 | CL18 | HEK-293T | C932(1.17) | LDD1609 | [9] |
| LDCM0406 | CL19 | HEK-293T | C932(1.00) | LDD1610 | [9] |
| LDCM0407 | CL2 | HEK-293T | C932(1.28) | LDD1611 | [9] |
| LDCM0408 | CL20 | HEK-293T | C932(1.32) | LDD1612 | [9] |
| LDCM0410 | CL22 | HEK-293T | C932(1.14) | LDD1614 | [9] |
| LDCM0411 | CL23 | HEK-293T | C932(1.07); C816(0.87) | LDD1615 | [9] |
| LDCM0412 | CL24 | HEK-293T | C932(0.98) | LDD1616 | [9] |
| LDCM0413 | CL25 | HEK-293T | C932(1.26); C404(1.13) | LDD1617 | [9] |
| LDCM0414 | CL26 | HEK-293T | C932(1.54) | LDD1618 | [9] |
| LDCM0415 | CL27 | HEK-293T | C932(1.06) | LDD1619 | [9] |
| LDCM0416 | CL28 | HEK-293T | C932(0.87) | LDD1620 | [9] |
| LDCM0417 | CL29 | HEK-293T | C932(1.07) | LDD1621 | [9] |
| LDCM0418 | CL3 | HEK-293T | C932(1.12) | LDD1622 | [9] |
| LDCM0419 | CL30 | HEK-293T | C932(1.02) | LDD1623 | [9] |
| LDCM0420 | CL31 | HEK-293T | C932(0.95) | LDD1624 | [9] |
| LDCM0421 | CL32 | HEK-293T | C932(1.15) | LDD1625 | [9] |
| LDCM0423 | CL34 | HEK-293T | C932(1.11) | LDD1627 | [9] |
| LDCM0424 | CL35 | HEK-293T | C932(1.15); C816(0.83) | LDD1628 | [9] |
| LDCM0425 | CL36 | HEK-293T | C932(1.00) | LDD1629 | [9] |
| LDCM0426 | CL37 | HEK-293T | C932(0.97); C404(1.14) | LDD1630 | [9] |
| LDCM0428 | CL39 | HEK-293T | C932(1.13) | LDD1632 | [9] |
| LDCM0429 | CL4 | HEK-293T | C932(0.92) | LDD1633 | [9] |
| LDCM0430 | CL40 | HEK-293T | C932(1.00) | LDD1634 | [9] |
| LDCM0431 | CL41 | HEK-293T | C932(1.17) | LDD1635 | [9] |
| LDCM0432 | CL42 | HEK-293T | C932(1.12) | LDD1636 | [9] |
| LDCM0433 | CL43 | HEK-293T | C932(0.83) | LDD1637 | [9] |
| LDCM0434 | CL44 | HEK-293T | C932(1.12) | LDD1638 | [9] |
| LDCM0436 | CL46 | HEK-293T | C932(1.11) | LDD1640 | [9] |
| LDCM0437 | CL47 | HEK-293T | C932(1.20); C816(1.05) | LDD1641 | [9] |
| LDCM0438 | CL48 | HEK-293T | C932(1.02) | LDD1642 | [9] |
| LDCM0439 | CL49 | HEK-293T | C932(1.12); C404(1.11) | LDD1643 | [9] |
| LDCM0440 | CL5 | HEK-293T | C932(1.11) | LDD1644 | [9] |
| LDCM0441 | CL50 | HEK-293T | C932(1.22) | LDD1645 | [9] |
| LDCM0443 | CL52 | HEK-293T | C932(0.98) | LDD1646 | [9] |
| LDCM0444 | CL53 | HEK-293T | C932(1.39) | LDD1647 | [9] |
| LDCM0445 | CL54 | HEK-293T | C932(1.20) | LDD1648 | [9] |
| LDCM0446 | CL55 | HEK-293T | C932(1.01) | LDD1649 | [9] |
| LDCM0447 | CL56 | HEK-293T | C932(1.18) | LDD1650 | [9] |
| LDCM0449 | CL58 | HEK-293T | C932(1.03) | LDD1652 | [9] |
| LDCM0450 | CL59 | HEK-293T | C932(1.14); C816(1.23) | LDD1653 | [9] |
| LDCM0451 | CL6 | HEK-293T | C932(1.25) | LDD1654 | [9] |
| LDCM0452 | CL60 | HEK-293T | C932(1.24) | LDD1655 | [9] |
| LDCM0453 | CL61 | HEK-293T | C932(1.01); C404(0.99) | LDD1656 | [9] |
| LDCM0454 | CL62 | HEK-293T | C932(1.32) | LDD1657 | [9] |
| LDCM0455 | CL63 | HEK-293T | C932(0.94) | LDD1658 | [9] |
| LDCM0456 | CL64 | HEK-293T | C932(1.02) | LDD1659 | [9] |
| LDCM0457 | CL65 | HEK-293T | C932(1.05) | LDD1660 | [9] |
| LDCM0458 | CL66 | HEK-293T | C932(0.85) | LDD1661 | [9] |
| LDCM0459 | CL67 | HEK-293T | C932(1.10) | LDD1662 | [9] |
| LDCM0460 | CL68 | HEK-293T | C932(1.28) | LDD1663 | [9] |
| LDCM0462 | CL7 | HEK-293T | C932(1.22) | LDD1665 | [9] |
| LDCM0463 | CL70 | HEK-293T | C932(1.00) | LDD1666 | [9] |
| LDCM0464 | CL71 | HEK-293T | C932(1.18); C816(0.79) | LDD1667 | [9] |
| LDCM0465 | CL72 | HEK-293T | C932(1.14) | LDD1668 | [9] |
| LDCM0466 | CL73 | HEK-293T | C932(1.19); C404(1.18) | LDD1669 | [9] |
| LDCM0467 | CL74 | HEK-293T | C932(1.21) | LDD1670 | [9] |
| LDCM0469 | CL76 | HEK-293T | C932(0.87) | LDD1672 | [9] |
| LDCM0470 | CL77 | HEK-293T | C932(1.21) | LDD1673 | [9] |
| LDCM0471 | CL78 | HEK-293T | C932(0.95) | LDD1674 | [9] |
| LDCM0472 | CL79 | HEK-293T | C932(1.08) | LDD1675 | [9] |
| LDCM0473 | CL8 | HEK-293T | C932(1.54) | LDD1676 | [9] |
| LDCM0474 | CL80 | HEK-293T | C932(1.14) | LDD1677 | [9] |
| LDCM0476 | CL82 | HEK-293T | C932(1.09) | LDD1679 | [9] |
| LDCM0477 | CL83 | HEK-293T | C932(1.17); C816(1.51) | LDD1680 | [9] |
| LDCM0478 | CL84 | HEK-293T | C932(1.23) | LDD1681 | [9] |
| LDCM0479 | CL85 | HEK-293T | C932(1.12); C404(1.13) | LDD1682 | [9] |
| LDCM0480 | CL86 | HEK-293T | C932(1.38) | LDD1683 | [9] |
| LDCM0481 | CL87 | HEK-293T | C932(1.15) | LDD1684 | [9] |
| LDCM0482 | CL88 | HEK-293T | C932(0.84) | LDD1685 | [9] |
| LDCM0483 | CL89 | HEK-293T | C932(1.17) | LDD1686 | [9] |
| LDCM0485 | CL90 | HEK-293T | C932(1.36) | LDD1688 | [9] |
| LDCM0486 | CL91 | HEK-293T | C932(1.12) | LDD1689 | [9] |
| LDCM0487 | CL92 | HEK-293T | C932(1.24) | LDD1690 | [9] |
| LDCM0489 | CL94 | HEK-293T | C932(1.01) | LDD1692 | [9] |
| LDCM0490 | CL95 | HEK-293T | C932(1.48); C816(1.15) | LDD1693 | [9] |
| LDCM0491 | CL96 | HEK-293T | C932(0.92) | LDD1694 | [9] |
| LDCM0492 | CL97 | HEK-293T | C932(0.93); C404(1.06) | LDD1695 | [9] |
| LDCM0493 | CL98 | HEK-293T | C932(1.09) | LDD1696 | [9] |
| LDCM0494 | CL99 | HEK-293T | C932(1.01) | LDD1697 | [9] |
| LDCM0495 | E2913 | HEK-293T | C932(1.00) | LDD1698 | [9] |
| LDCM0468 | Fragment33 | HEK-293T | C932(1.16) | LDD1671 | [9] |
| LDCM0427 | Fragment51 | HEK-293T | C932(1.15) | LDD1631 | [9] |
| LDCM0022 | KB02 | A2058 | C686(3.44); C928(3.88) | LDD2253 | [3] |
| LDCM0023 | KB03 | MDA-MB-231 | C656(1.00) | LDD1701 | [10] |
| LDCM0024 | KB05 | G361 | C962(15.26) | LDD3311 | [3] |
| LDCM0090 | Rapamycin | JHH-7 | 8.80 | LDD0213 | [8] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| NAD-dependent protein deacetylase sirtuin-1 (SIRT1) | Sirtuin family | Q96EB6 | |||
| CREB-binding protein (CREBBP) | . | Q92793 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Ataxin-1 (ATXN1) | ATXN1 family | P54253 | |||
| Mediator of RNA polymerase II transcription subunit 15 (MED15) | Mediator complex subunit 15 family | Q96RN5 | |||
References
