Details of the Target
General Information of Target
| Target ID | LDTP08874 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mitochondrial intermembrane space import and assembly protein 40 (CHCHD4) | |||||
| Gene Name | CHCHD4 | |||||
| Gene ID | 131474 | |||||
| Synonyms |
MIA40; Mitochondrial intermembrane space import and assembly protein 40; Coiled-coil-helix-coiled-coil-helix domain-containing protein 4 |
|||||
| 3D Structure | ||||||
| Sequence |
MSYCRQEGKDRIIFVTKEDHETPSSAELVADDPNDPYEEHGLILPNGNINWNCPCLGGMA
SGPCGEQFKSAFSCFHYSTEEIKGSDCVDQFRAMQECMQKYPDLYPQEDEDEEEEREKKP AEQAEETAPIEATATKEEEGSS |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Mitochondrion intermembrane space
|
|||||
| Function |
Central component of a redox-sensitive mitochondrial intermembrane space import machinery which is required for the biogenesis of respiratory chain complexes. Functions as chaperone and catalyzes the formation of disulfide bonds in substrate proteins, such as COX17, COX19, MICU1 and COA7. Required for the import and folding of small cysteine-containing proteins (small Tim) in the mitochondrial intermembrane space (IMS). Required for the import of COA7 in the IMS. Precursor proteins to be imported into the IMS are translocated in their reduced form into the mitochondria. The oxidized form of CHCHD4/MIA40 forms a transient intermolecular disulfide bridge with the reduced precursor protein, resulting in oxidation of the precursor protein that now contains an intramolecular disulfide bond and is able to undergo folding in the IMS. Reduced CHCHD4/MIA40 is then reoxidized by GFER/ERV1 via a disulfide relay system. Mediates formation of disulfide bond in MICU1 in the IMS, promoting formation of the MICU1-MICU2 heterodimer that regulates mitochondrial calcium uptake.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FBP2 Probe Info |
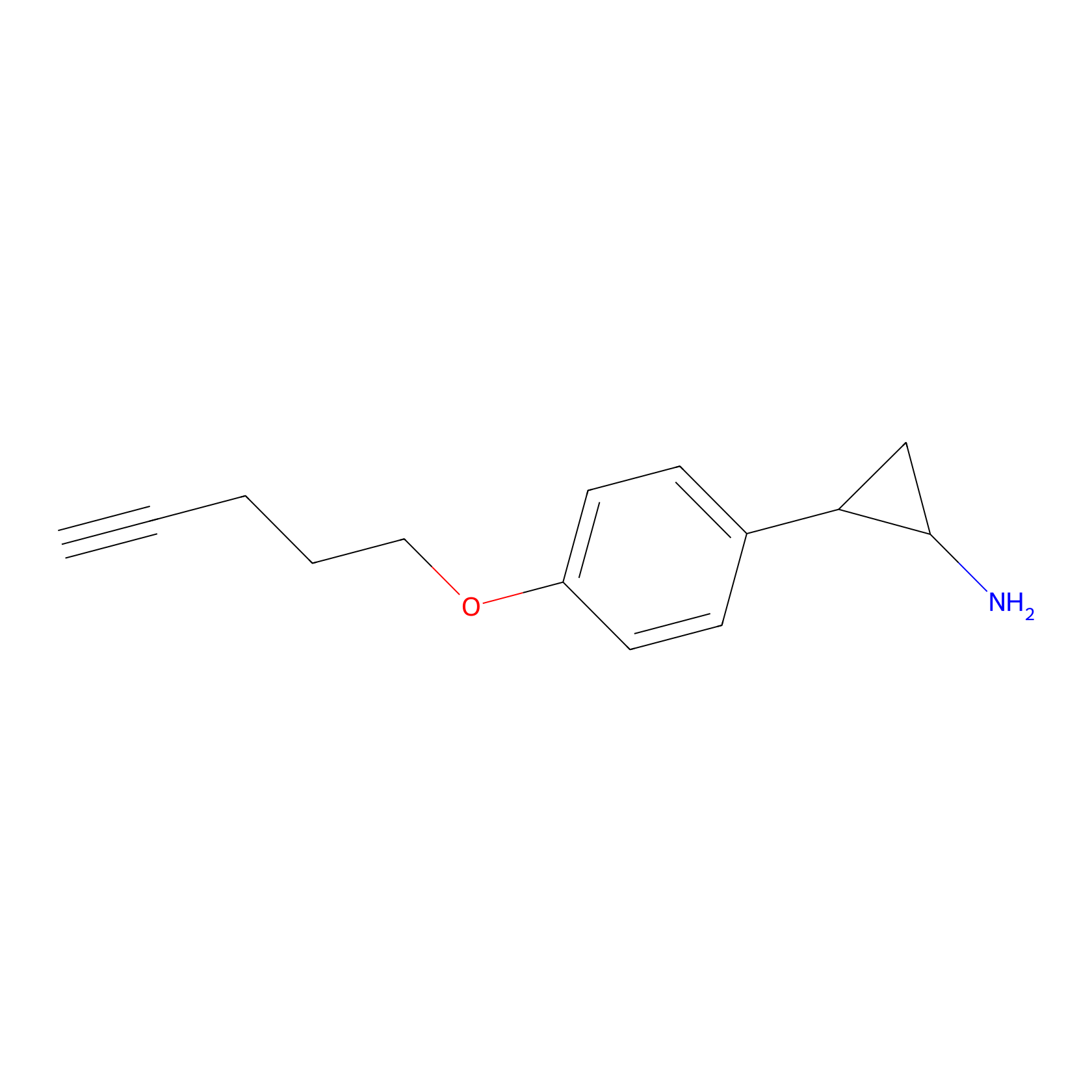 |
3.26 | LDD0323 | [1] | |
|
Probe 1 Probe Info |
 |
Y101(12.62); Y105(11.12) | LDD3495 | [2] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [3] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [4] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C97(0.00); C74(0.00) | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
C97(0.00); C87(0.00); C74(0.00) | LDD0036 | [5] | |
|
Lodoacetamide azide Probe Info |
 |
C87(0.00); C74(0.00) | LDD0037 | [5] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [6] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [7] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [8] | |
|
NAIA_5 Probe Info |
 |
C97(0.00); C87(0.00); C74(0.00); C4(0.00) | LDD2223 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C265 Probe Info |
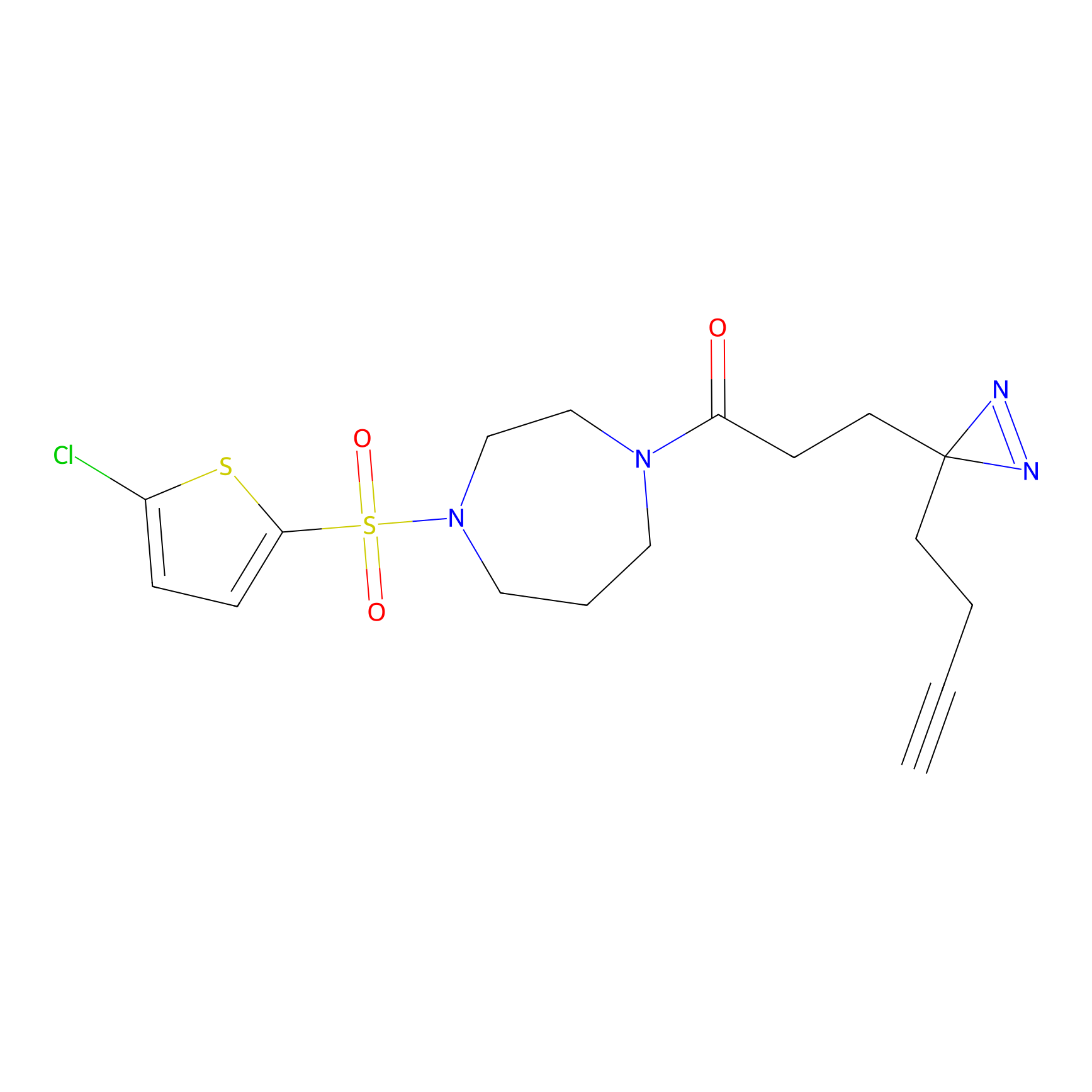 |
17.88 | LDD1936 | [10] | |
|
C342 Probe Info |
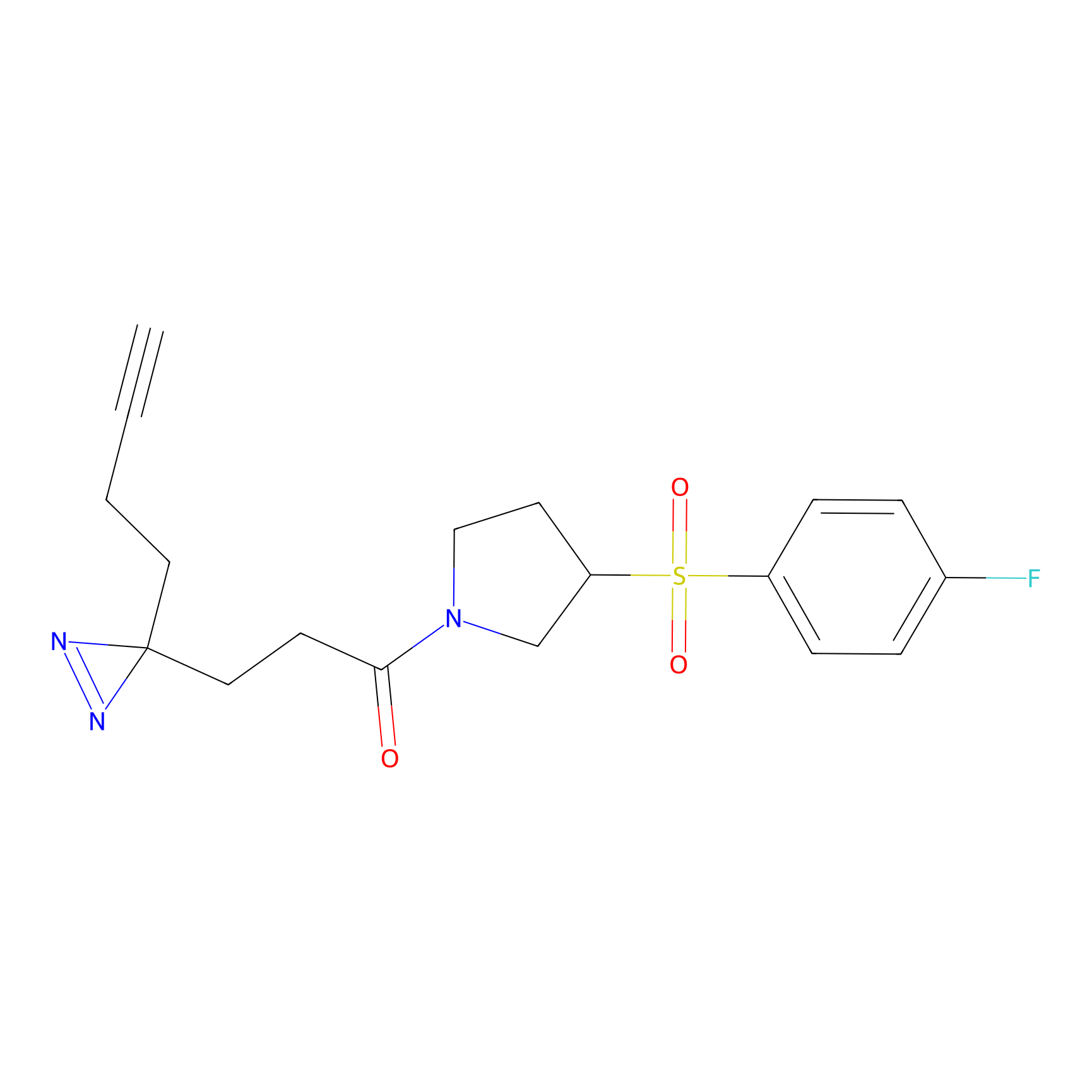 |
6.32 | LDD2004 | [10] | |
|
C343 Probe Info |
 |
11.31 | LDD2005 | [10] | |
|
C431 Probe Info |
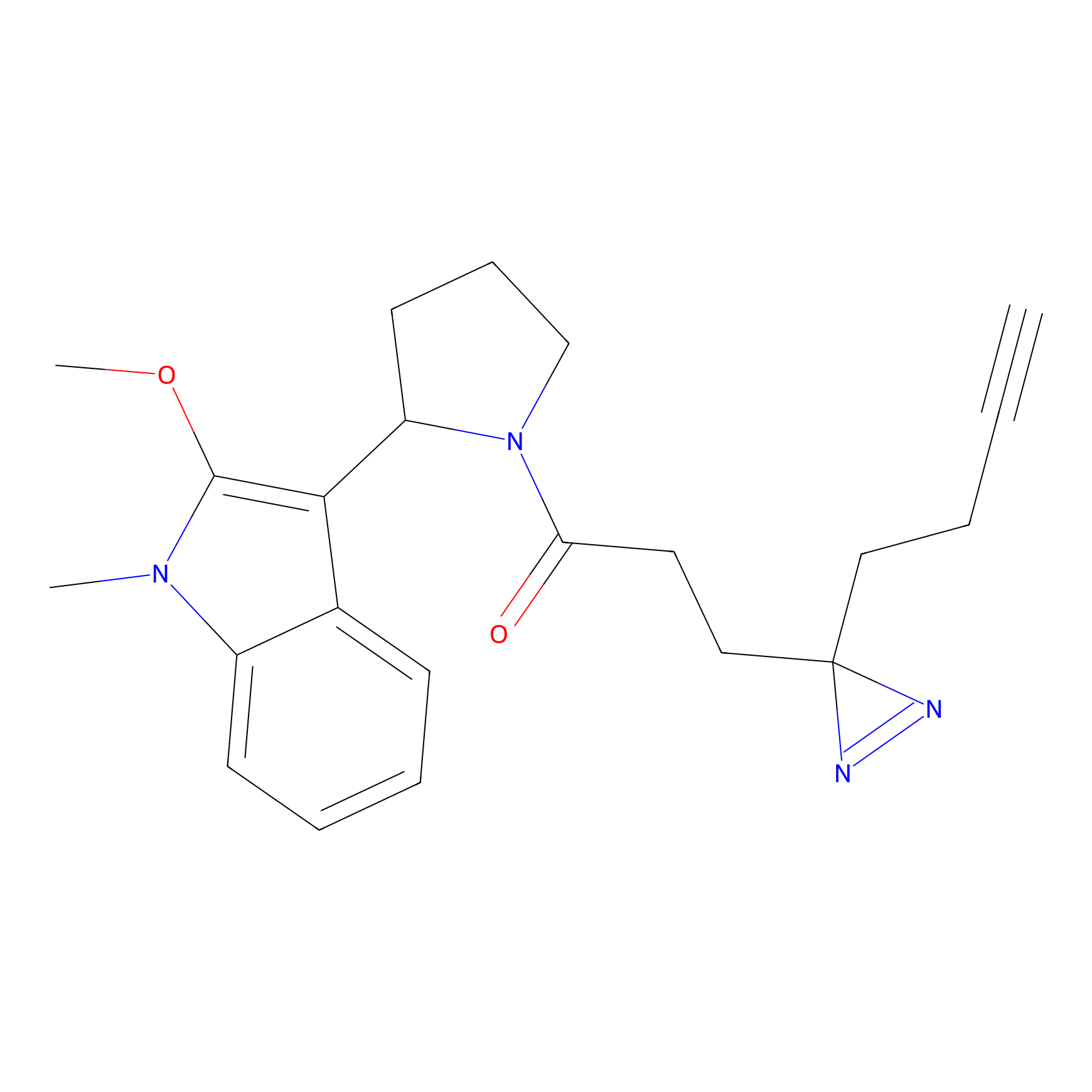 |
13.83 | LDD2086 | [10] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
