Details of the Target
General Information of Target
| Target ID | LDTP07819 | |||||
|---|---|---|---|---|---|---|
| Target Name | Rho family-interacting cell polarization regulator 1 (RIPOR1) | |||||
| Gene Name | RIPOR1 | |||||
| Gene ID | 79567 | |||||
| Synonyms |
FAM65A; KIAA1930; Rho family-interacting cell polarization regulator 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MMSLSVRPQRRLLSARVNRSQSFAGVLGSHERGPSLSFRSFPVFSPPGPPRKPPALSRVS
RMFSVAHPAAKVPQPERLDLVYTALKRGLTAYLEVHQQEQEKLQGQIRESKRNSRLGFLY DLDKQVKSIERFLRRLEFHASKIDELYEAYCVQRRLRDGAYNMVRAYTTGSPGSREARDS LAEATRGHREYTESMCLLESELEAQLGEFHLRMKGLAGFARLCVGDQYEICMKYGRQRWK LRGRIEGSGKQVWDSEETIFLPLLTEFLSIKVTELKGLANHVVVGSVSCETKDLFAALPQ VVAVDINDLGTIKLSLEVTWSPFDKDDQPSAASSVNKASTVTKRFSTYSQSPPDTPSLRE QAFYNMLRRQEELENGTAWSLSSESSDDSSSPQLSGTARHSPAPRPLVQQPEPLPIQVAF RRPETPSSGPLDEEGAVAPVLANGHAPYSRTLSHISEASVDAALAEASVEAVGPESLAWG PSPPTHPAPTHGEHPSPVPPALDPGHSATSSTLGTTGSVPTSTDPAPSAHLDSVHKSTDS GPSELPGPTHTTTGSTYSAITTTHSAPSPLTHTTTGSTHKPIISTLTTTGPTLNIIGPVQ TTTSPTHTMPSPTHTTASPTHTSTSPTHTPTSPTHKTSMSPPTTTSPTPSGMGLVQTATS PTHPTTSPTHPTTSPILINVSPSTSLELATLSSPSKHSDPTLPGTDSLPCSPPVSNSYTQ ADPMAPRTPHPSPAHSSRKPLTSPAPDPSESTVQSLSPTPSPPTPAPQHSDLCLAMAVQT PVPTAAGGSGDRSLEEALGALMAALDDYRGQFPELQGLEQEVTRLESLLMQRQGLTRSRA SSLSITVEHALESFSFLNEDEDEDNDVPGDRPPSSPEAGAEDSIDSPSARPLSTGCPALD AALVRHLYHCSRLLLKLGTFGPLRCQEAWALERLLREARVLEAVCEFSRRWEIPASSAQE VVQFSASRPGFLTFWDQCTERLSCFLCPVERVLLTFCNQYGARLSLRQPGLAEAVCVKFL EDALGQKLPRRPQPGPGEQLTVFQFWSFVETLDSPTMEAYVTETAEEVLLVRNLNSDDQA VVLKALRLAPEGRLRRDGLRALSSLLVHGNNKVMAAVSTQLRSLSLGPTFRERALLCFLD QLEDEDVQTRVAGCLALGCIKAPEGIEPLVYLCQTDTEAVREAARQSLQQCGEEGQSAHR RLEESLDALPRIFGPGSMASTAF |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
RIPOR family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Downstream effector protein for Rho-type small GTPases that plays a role in cell polarity and directional migration. Acts as an adapter protein, linking active Rho proteins to STK24 and STK26 kinases, and hence positively regulates Golgi reorientation in polarized cell migration upon Rho activation. Involved in the subcellular relocation of STK26 from the Golgi to cytoplasm punctae in a Rho- and PDCD10-dependent manner upon serum stimulation.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K1084(8.33); K343(4.21); K86(10.00) | LDD0277 | [2] | |
|
OPA-S-S-alkyne Probe Info |
 |
K71(5.33) | LDD3494 | [3] | |
|
BTD Probe Info |
 |
C1191(1.36) | LDD1700 | [4] | |
|
Johansson_61 Probe Info |
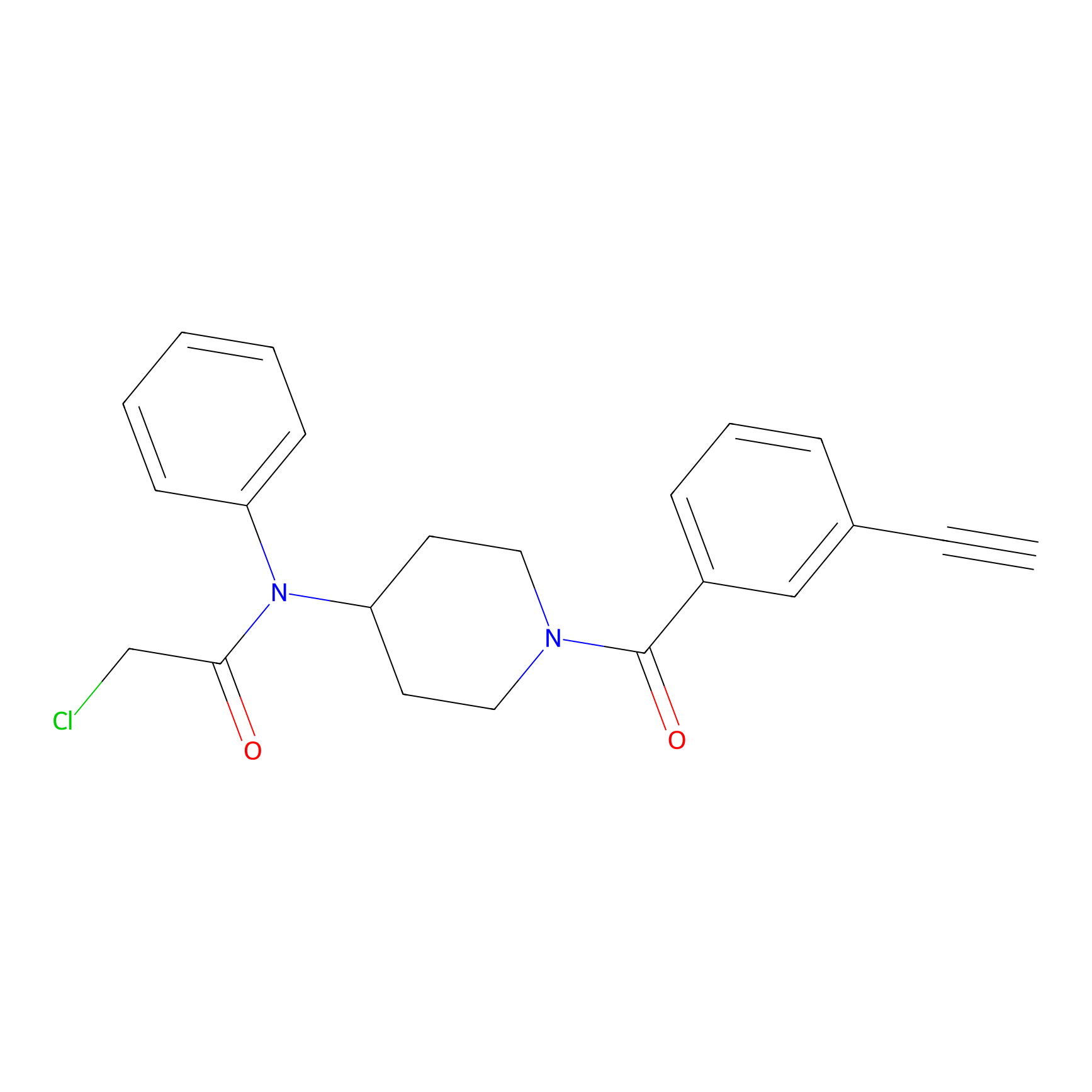 |
_(4.25) | LDD1487 | [5] | |
|
AHL-Pu-1 Probe Info |
 |
C997(2.33) | LDD0170 | [6] | |
|
DBIA Probe Info |
 |
C925(34.00) | LDD0209 | [7] | |
|
IPM Probe Info |
 |
C151(1.50) | LDD1701 | [4] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [8] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2225 | [9] | |
|
TFBX Probe Info |
 |
C910(0.00); C984(0.00) | LDD0148 | [10] | |
|
AOyne Probe Info |
 |
6.80 | LDD0443 | [11] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C910(0.62) | LDD2142 | [4] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C945(1.25) | LDD2117 | [4] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C1191(1.22) | LDD2152 | [4] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C1191(0.61) | LDD2132 | [4] |
| LDCM0025 | 4SU-RNA | DM93 | C997(2.33) | LDD0170 | [6] |
| LDCM0214 | AC1 | HEK-293T | C1137(1.02) | LDD1507 | [12] |
| LDCM0215 | AC10 | HEK-293T | C1137(1.44) | LDD1508 | [12] |
| LDCM0226 | AC11 | HEK-293T | C1137(1.02); C896(0.94); C925(0.96) | LDD1509 | [12] |
| LDCM0237 | AC12 | HEK-293T | C1137(1.24); C896(1.16) | LDD1510 | [12] |
| LDCM0259 | AC14 | HEK-293T | C1137(0.88); C896(1.01); C925(1.29) | LDD1512 | [12] |
| LDCM0270 | AC15 | HEK-293T | C896(0.97); C1173(0.89) | LDD1513 | [12] |
| LDCM0276 | AC17 | HEK-293T | C1137(0.92) | LDD1515 | [12] |
| LDCM0277 | AC18 | HEK-293T | C1137(1.31) | LDD1516 | [12] |
| LDCM0278 | AC19 | HEK-293T | C1137(0.78); C896(0.85); C925(1.24) | LDD1517 | [12] |
| LDCM0279 | AC2 | HEK-293T | C1137(1.25) | LDD1518 | [12] |
| LDCM0280 | AC20 | HEK-293T | C1137(1.24); C896(1.05) | LDD1519 | [12] |
| LDCM0281 | AC21 | HEK-293T | C1137(0.88); C896(1.11) | LDD1520 | [12] |
| LDCM0282 | AC22 | HEK-293T | C1137(1.10); C896(1.05); C925(1.16) | LDD1521 | [12] |
| LDCM0283 | AC23 | HEK-293T | C896(1.00); C1173(0.88) | LDD1522 | [12] |
| LDCM0285 | AC25 | HEK-293T | C1137(1.12) | LDD1524 | [12] |
| LDCM0286 | AC26 | HEK-293T | C1137(1.39) | LDD1525 | [12] |
| LDCM0287 | AC27 | HEK-293T | C1137(1.27); C896(1.14); C925(1.17) | LDD1526 | [12] |
| LDCM0288 | AC28 | HEK-293T | C1137(1.33); C896(1.08) | LDD1527 | [12] |
| LDCM0289 | AC29 | HEK-293T | C1137(0.94); C896(1.09) | LDD1528 | [12] |
| LDCM0290 | AC3 | HEK-293T | C1137(0.79); C896(1.02); C925(0.93) | LDD1529 | [12] |
| LDCM0291 | AC30 | HEK-293T | C1137(0.91); C896(0.83); C925(1.00) | LDD1530 | [12] |
| LDCM0292 | AC31 | HEK-293T | C896(0.93); C1173(0.68) | LDD1531 | [12] |
| LDCM0294 | AC33 | HEK-293T | C1137(0.85) | LDD1533 | [12] |
| LDCM0295 | AC34 | HEK-293T | C1137(0.93) | LDD1534 | [12] |
| LDCM0296 | AC35 | HEK-293T | C1137(1.04); C896(0.98); C925(0.99) | LDD1535 | [12] |
| LDCM0297 | AC36 | HEK-293T | C1137(1.02); C896(1.01) | LDD1536 | [12] |
| LDCM0298 | AC37 | HEK-293T | C1137(0.88); C896(1.12) | LDD1537 | [12] |
| LDCM0299 | AC38 | HEK-293T | C1137(0.90); C896(1.21); C925(1.16) | LDD1538 | [12] |
| LDCM0300 | AC39 | HEK-293T | C896(0.96); C1173(0.86) | LDD1539 | [12] |
| LDCM0301 | AC4 | HEK-293T | C1137(1.08); C896(1.02) | LDD1540 | [12] |
| LDCM0303 | AC41 | HEK-293T | C1137(0.98) | LDD1542 | [12] |
| LDCM0304 | AC42 | HEK-293T | C1137(0.96) | LDD1543 | [12] |
| LDCM0305 | AC43 | HEK-293T | C1137(0.82); C896(1.25); C925(1.17) | LDD1544 | [12] |
| LDCM0306 | AC44 | HEK-293T | C1137(1.21); C896(1.02) | LDD1545 | [12] |
| LDCM0307 | AC45 | HEK-293T | C1137(0.98); C896(1.22) | LDD1546 | [12] |
| LDCM0308 | AC46 | HEK-293T | C1137(0.88); C896(1.02); C925(1.19) | LDD1547 | [12] |
| LDCM0309 | AC47 | HEK-293T | C896(0.95); C1173(0.95) | LDD1548 | [12] |
| LDCM0311 | AC49 | HEK-293T | C1137(1.16) | LDD1550 | [12] |
| LDCM0312 | AC5 | HEK-293T | C1137(0.88); C896(1.15) | LDD1551 | [12] |
| LDCM0313 | AC50 | HEK-293T | C1137(1.20) | LDD1552 | [12] |
| LDCM0314 | AC51 | HEK-293T | C1137(0.96); C896(0.88); C925(1.07) | LDD1553 | [12] |
| LDCM0315 | AC52 | HEK-293T | C1137(1.05); C896(1.07) | LDD1554 | [12] |
| LDCM0316 | AC53 | HEK-293T | C1137(0.92); C896(1.12) | LDD1555 | [12] |
| LDCM0317 | AC54 | HEK-293T | C1137(1.11); C896(1.01); C925(1.33) | LDD1556 | [12] |
| LDCM0318 | AC55 | HEK-293T | C896(0.96); C1173(0.87) | LDD1557 | [12] |
| LDCM0320 | AC57 | HEK-293T | C1137(1.07) | LDD1559 | [12] |
| LDCM0321 | AC58 | HEK-293T | C1137(1.45) | LDD1560 | [12] |
| LDCM0322 | AC59 | HEK-293T | C1137(1.38); C896(0.88); C925(1.08) | LDD1561 | [12] |
| LDCM0323 | AC6 | HEK-293T | C1137(0.99); C896(1.13); C925(1.14) | LDD1562 | [12] |
| LDCM0324 | AC60 | HEK-293T | C1137(1.11); C896(1.10) | LDD1563 | [12] |
| LDCM0325 | AC61 | HEK-293T | C1137(0.86); C896(1.04) | LDD1564 | [12] |
| LDCM0326 | AC62 | HEK-293T | C1137(0.89); C896(1.21); C925(1.26) | LDD1565 | [12] |
| LDCM0327 | AC63 | HEK-293T | C896(0.89); C1173(0.90) | LDD1566 | [12] |
| LDCM0334 | AC7 | HEK-293T | C896(0.99); C1173(0.90) | LDD1568 | [12] |
| LDCM0545 | Acetamide | MDA-MB-231 | C1191(0.92) | LDD2138 | [4] |
| LDCM0248 | AKOS034007472 | HEK-293T | C1137(0.74); C896(1.10) | LDD1511 | [12] |
| LDCM0356 | AKOS034007680 | HEK-293T | C1137(1.02) | LDD1570 | [12] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C1191(0.48) | LDD2091 | [4] |
| LDCM0632 | CL-Sc | Hep-G2 | C997(1.05) | LDD2227 | [9] |
| LDCM0367 | CL1 | HEK-293T | C1137(1.04); C896(1.28) | LDD1571 | [12] |
| LDCM0368 | CL10 | HEK-293T | C1137(0.92); C896(1.53); C925(1.39) | LDD1572 | [12] |
| LDCM0370 | CL101 | HEK-293T | C1137(1.06); C896(1.25) | LDD1574 | [12] |
| LDCM0371 | CL102 | HEK-293T | C896(0.96) | LDD1575 | [12] |
| LDCM0372 | CL103 | HEK-293T | C1137(0.93) | LDD1576 | [12] |
| LDCM0374 | CL105 | HEK-293T | C1137(0.92); C896(1.31) | LDD1578 | [12] |
| LDCM0375 | CL106 | HEK-293T | C896(0.87) | LDD1579 | [12] |
| LDCM0376 | CL107 | HEK-293T | C1137(1.07) | LDD1580 | [12] |
| LDCM0378 | CL109 | HEK-293T | C1137(0.79); C896(1.11) | LDD1582 | [12] |
| LDCM0379 | CL11 | HEK-293T | C896(1.23); C1173(1.33) | LDD1583 | [12] |
| LDCM0380 | CL110 | HEK-293T | C896(0.86) | LDD1584 | [12] |
| LDCM0381 | CL111 | HEK-293T | C1137(1.38) | LDD1585 | [12] |
| LDCM0383 | CL113 | HEK-293T | C1137(0.94); C896(1.22) | LDD1587 | [12] |
| LDCM0384 | CL114 | HEK-293T | C896(1.04) | LDD1588 | [12] |
| LDCM0385 | CL115 | HEK-293T | C1137(1.13) | LDD1589 | [12] |
| LDCM0387 | CL117 | HEK-293T | C1137(0.84); C896(1.17) | LDD1591 | [12] |
| LDCM0388 | CL118 | HEK-293T | C896(0.88) | LDD1592 | [12] |
| LDCM0389 | CL119 | HEK-293T | C1137(1.05) | LDD1593 | [12] |
| LDCM0392 | CL121 | HEK-293T | C1137(1.39); C896(1.18) | LDD1596 | [12] |
| LDCM0393 | CL122 | HEK-293T | C896(1.16) | LDD1597 | [12] |
| LDCM0394 | CL123 | HEK-293T | C1137(0.82) | LDD1598 | [12] |
| LDCM0396 | CL125 | HEK-293T | C1137(1.04); C896(1.25) | LDD1600 | [12] |
| LDCM0397 | CL126 | HEK-293T | C896(0.89) | LDD1601 | [12] |
| LDCM0398 | CL127 | HEK-293T | C1137(0.79) | LDD1602 | [12] |
| LDCM0400 | CL13 | HEK-293T | C1137(0.97); C896(1.42) | LDD1604 | [12] |
| LDCM0401 | CL14 | HEK-293T | C896(0.98) | LDD1605 | [12] |
| LDCM0402 | CL15 | HEK-293T | C1137(0.77) | LDD1606 | [12] |
| LDCM0404 | CL17 | HEK-293T | C1137(1.07) | LDD1608 | [12] |
| LDCM0405 | CL18 | HEK-293T | C1137(1.25) | LDD1609 | [12] |
| LDCM0406 | CL19 | HEK-293T | C1137(1.06); C896(1.11); C925(1.11) | LDD1610 | [12] |
| LDCM0407 | CL2 | HEK-293T | C896(0.85) | LDD1611 | [12] |
| LDCM0408 | CL20 | HEK-293T | C1137(0.99); C896(1.34) | LDD1612 | [12] |
| LDCM0409 | CL21 | HEK-293T | C1137(0.77); C896(1.27) | LDD1613 | [12] |
| LDCM0410 | CL22 | HEK-293T | C1137(0.92); C896(1.35); C925(1.19) | LDD1614 | [12] |
| LDCM0411 | CL23 | HEK-293T | C896(1.09); C1173(0.99) | LDD1615 | [12] |
| LDCM0413 | CL25 | HEK-293T | C1137(0.60); C896(1.18) | LDD1617 | [12] |
| LDCM0414 | CL26 | HEK-293T | C896(0.90) | LDD1618 | [12] |
| LDCM0415 | CL27 | HEK-293T | C1137(1.07) | LDD1619 | [12] |
| LDCM0417 | CL29 | HEK-293T | C1137(0.94) | LDD1621 | [12] |
| LDCM0418 | CL3 | HEK-293T | C1137(0.87) | LDD1622 | [12] |
| LDCM0419 | CL30 | HEK-293T | C1137(1.15) | LDD1623 | [12] |
| LDCM0420 | CL31 | HEK-293T | C1137(0.97); C896(1.10); C925(1.10) | LDD1624 | [12] |
| LDCM0421 | CL32 | HEK-293T | C1137(1.09); C896(1.32) | LDD1625 | [12] |
| LDCM0422 | CL33 | HEK-293T | C1137(0.83); C896(1.36) | LDD1626 | [12] |
| LDCM0423 | CL34 | HEK-293T | C1137(0.91); C896(1.18); C925(1.12) | LDD1627 | [12] |
| LDCM0424 | CL35 | HEK-293T | C896(1.03); C1173(0.86) | LDD1628 | [12] |
| LDCM0426 | CL37 | HEK-293T | C1137(1.15); C896(1.18) | LDD1630 | [12] |
| LDCM0428 | CL39 | HEK-293T | C1137(1.11) | LDD1632 | [12] |
| LDCM0431 | CL41 | HEK-293T | C1137(0.92) | LDD1635 | [12] |
| LDCM0432 | CL42 | HEK-293T | C1137(1.13) | LDD1636 | [12] |
| LDCM0433 | CL43 | HEK-293T | C1137(0.72); C896(1.13); C925(1.06) | LDD1637 | [12] |
| LDCM0434 | CL44 | HEK-293T | C1137(1.01); C896(1.07) | LDD1638 | [12] |
| LDCM0435 | CL45 | HEK-293T | C1137(1.17); C896(1.18) | LDD1639 | [12] |
| LDCM0436 | CL46 | HEK-293T | C1137(0.99); C896(1.26); C925(1.41) | LDD1640 | [12] |
| LDCM0437 | CL47 | HEK-293T | C896(1.08); C1173(1.13) | LDD1641 | [12] |
| LDCM0439 | CL49 | HEK-293T | C1137(1.15); C896(1.19) | LDD1643 | [12] |
| LDCM0440 | CL5 | HEK-293T | C1137(0.98) | LDD1644 | [12] |
| LDCM0441 | CL50 | HEK-293T | C896(0.88) | LDD1645 | [12] |
| LDCM0444 | CL53 | HEK-293T | C1137(0.90) | LDD1647 | [12] |
| LDCM0445 | CL54 | HEK-293T | C1137(0.89) | LDD1648 | [12] |
| LDCM0446 | CL55 | HEK-293T | C1137(1.10); C896(1.01); C925(1.06) | LDD1649 | [12] |
| LDCM0447 | CL56 | HEK-293T | C1137(1.05); C896(1.46) | LDD1650 | [12] |
| LDCM0448 | CL57 | HEK-293T | C1137(0.88); C896(1.25) | LDD1651 | [12] |
| LDCM0449 | CL58 | HEK-293T | C1137(1.05); C896(1.26); C925(1.30) | LDD1652 | [12] |
| LDCM0450 | CL59 | HEK-293T | C896(1.17); C1173(1.00) | LDD1653 | [12] |
| LDCM0451 | CL6 | HEK-293T | C1137(0.87) | LDD1654 | [12] |
| LDCM0453 | CL61 | HEK-293T | C1137(1.09); C896(1.31) | LDD1656 | [12] |
| LDCM0454 | CL62 | HEK-293T | C896(0.78) | LDD1657 | [12] |
| LDCM0455 | CL63 | HEK-293T | C1137(0.97) | LDD1658 | [12] |
| LDCM0457 | CL65 | HEK-293T | C1137(1.11) | LDD1660 | [12] |
| LDCM0458 | CL66 | HEK-293T | C1137(1.20) | LDD1661 | [12] |
| LDCM0459 | CL67 | HEK-293T | C1137(1.26); C896(1.20); C925(1.17) | LDD1662 | [12] |
| LDCM0460 | CL68 | HEK-293T | C1137(1.19); C896(1.33) | LDD1663 | [12] |
| LDCM0461 | CL69 | HEK-293T | C1137(0.87); C896(1.19) | LDD1664 | [12] |
| LDCM0462 | CL7 | HEK-293T | C1137(0.94); C896(1.04); C925(1.22) | LDD1665 | [12] |
| LDCM0463 | CL70 | HEK-293T | C1137(1.06); C896(1.07); C925(1.21) | LDD1666 | [12] |
| LDCM0464 | CL71 | HEK-293T | C896(1.05); C1173(0.94) | LDD1667 | [12] |
| LDCM0466 | CL73 | HEK-293T | C1137(0.80); C896(1.26) | LDD1669 | [12] |
| LDCM0467 | CL74 | HEK-293T | C896(0.98) | LDD1670 | [12] |
| LDCM0470 | CL77 | HEK-293T | C1137(0.96) | LDD1673 | [12] |
| LDCM0471 | CL78 | HEK-293T | C1137(1.21) | LDD1674 | [12] |
| LDCM0472 | CL79 | HEK-293T | C1137(1.19); C896(1.07); C925(1.00) | LDD1675 | [12] |
| LDCM0473 | CL8 | HEK-293T | C1137(1.13); C896(1.85) | LDD1676 | [12] |
| LDCM0474 | CL80 | HEK-293T | C1137(1.09); C896(1.25) | LDD1677 | [12] |
| LDCM0475 | CL81 | HEK-293T | C1137(1.13); C896(1.17) | LDD1678 | [12] |
| LDCM0476 | CL82 | HEK-293T | C1137(1.12); C896(1.13); C925(1.47) | LDD1679 | [12] |
| LDCM0477 | CL83 | HEK-293T | C896(0.88); C1173(1.11) | LDD1680 | [12] |
| LDCM0479 | CL85 | HEK-293T | C1137(0.95); C896(1.28) | LDD1682 | [12] |
| LDCM0480 | CL86 | HEK-293T | C896(1.00) | LDD1683 | [12] |
| LDCM0481 | CL87 | HEK-293T | C1137(1.05) | LDD1684 | [12] |
| LDCM0483 | CL89 | HEK-293T | C1137(0.93) | LDD1686 | [12] |
| LDCM0484 | CL9 | HEK-293T | C1137(0.87); C896(1.15) | LDD1687 | [12] |
| LDCM0485 | CL90 | HEK-293T | C1137(1.04) | LDD1688 | [12] |
| LDCM0486 | CL91 | HEK-293T | C1137(1.27); C896(0.95); C925(0.98) | LDD1689 | [12] |
| LDCM0487 | CL92 | HEK-293T | C1137(1.10); C896(1.29) | LDD1690 | [12] |
| LDCM0488 | CL93 | HEK-293T | C1137(0.82); C896(1.25) | LDD1691 | [12] |
| LDCM0489 | CL94 | HEK-293T | C1137(1.30); C896(1.21); C925(1.16) | LDD1692 | [12] |
| LDCM0490 | CL95 | HEK-293T | C896(0.89); C1173(1.17) | LDD1693 | [12] |
| LDCM0492 | CL97 | HEK-293T | C1137(0.84); C896(1.20) | LDD1695 | [12] |
| LDCM0493 | CL98 | HEK-293T | C896(0.98) | LDD1696 | [12] |
| LDCM0494 | CL99 | HEK-293T | C1137(0.93) | LDD1697 | [12] |
| LDCM0495 | E2913 | HEK-293T | C1137(1.10) | LDD1698 | [12] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C987(3.32); C151(1.71); C1159(0.98) | LDD1702 | [4] |
| LDCM0468 | Fragment33 | HEK-293T | C1137(0.94) | LDD1671 | [12] |
| LDCM0427 | Fragment51 | HEK-293T | C896(1.15) | LDD1631 | [12] |
| LDCM0615 | Fragment63-R | Jurkat | _(4.25) | LDD1487 | [5] |
| LDCM0022 | KB02 | HEK-293T | C1137(0.79) | LDD1492 | [12] |
| LDCM0023 | KB03 | Jurkat | C925(34.00) | LDD0209 | [7] |
| LDCM0024 | KB05 | UACC257 | C1032(3.66); C997(2.51) | LDD3325 | [13] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C1191(0.61) | LDD2121 | [4] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C1191(0.47) | LDD2089 | [4] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C1191(0.66) | LDD2092 | [4] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C1191(0.84) | LDD2093 | [4] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C1191(0.82) | LDD2101 | [4] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C1191(1.04) | LDD2105 | [4] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C910(0.28) | LDD2108 | [4] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C1191(1.18) | LDD2109 | [4] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C1191(0.52) | LDD2115 | [4] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C945(1.26) | LDD2123 | [4] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C1191(0.69) | LDD2125 | [4] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C945(1.51); C1191(0.88) | LDD2129 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C945(1.40) | LDD2136 | [4] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C1191(1.36) | LDD1700 | [4] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C1191(0.49) | LDD2140 | [4] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C945(1.90) | LDD2144 | [4] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C1191(0.99) | LDD2146 | [4] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C1191(0.43) | LDD2150 | [4] |
References
