Details of the Target
General Information of Target
| Target ID | LDTP05814 | |||||
|---|---|---|---|---|---|---|
| Target Name | Battenin (CLN3) | |||||
| Gene Name | CLN3 | |||||
| Gene ID | 1201 | |||||
| Synonyms |
BTS; Battenin; Batten disease protein; Protein CLN3 |
|||||
| 3D Structure | ||||||
| Sequence |
MGGCAGSRRRFSDSEGEETVPEPRLPLLDHQGAHWKNAVGFWLLGLCNNFSYVVMLSAAH
DILSHKRTSGNQSHVDPGPTPIPHNSSSRFDCNSVSTAAVLLADILPTLVIKLLAPLGLH LLPYSPRVLVSGICAAGSFVLVAFSHSVGTSLCGVVFASISSGLGEVTFLSLTAFYPRAV ISWWSSGTGGAGLLGALSYLGLTQAGLSPQQTLLSMLGIPALLLASYFLLLTSPEAQDPG GEEEAESAARQPLIRTEAPESKPGSSSSLSLRERWTVFKGLLWYIVPLVVVYFAEYFINQ GLFELLFFWNTSLSHAQQYRWYQMLYQAGVFASRSSLRCCRIRFTWALALLQCLNLVFLL ADVWFGFLPSIYLVFLIILYEGLLGGAAYVNTFHNIALETSDEHREFAMAATCISDTLGI SLSGLLALPLHDFLCQLS |
|||||
| Target Type |
Clinical trial
|
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Battenin family
|
|||||
| Subcellular location |
Lysosome membrane
|
|||||
| Function |
Mediates microtubule-dependent, anterograde transport connecting the Golgi network, endosomes, autophagosomes, lysosomes and plasma membrane, and participates in several cellular processes such as regulation of lysosomal pH, lysosome protein degradation, receptor-mediated endocytosis, autophagy, transport of proteins and lipids from the TGN, apoptosis and synaptic transmission . Facilitates the proteins transport from trans-Golgi network (TGN)-to other membrane compartments such as transport of microdomain-associated proteins to the plasma membrane, IGF2R transport to the lysosome where it regulates the CTSD release leading to regulation of CTSD maturation and thereby APP intracellular processing. Moreover regulates CTSD activity in response to osmotic stress. Also binds galactosylceramide and transports it from the trans Golgi to the rafts, which may have immediate and downstream effects on cell survival by modulating ceramide synthesis. At the plasma membrane, regulates actin-dependent events including filopodia formation, cell migration, and pinocytosis through ARF1-CDC42 pathway and also the cytoskeleton organization through interaction with MYH10 and fodrin leading to the regulation of the plasma membrane association of Na+, K+ ATPase complex. Regulates synaptic transmission in the amygdala, hippocampus, and cerebellum through regulation of synaptic vesicles density and their proximity to active zones leading to modulation of short-term plasticity and age-dependent anxious behavior, learning and memory. Regulates autophagic vacuoles (AVs) maturation by modulating the trafficking between endocytic and autophagolysosomal/lysosomal compartments, which involves vesicle fusion leading to regulation of degradation process. Participates also in cellular homeostasis of compounds such as, water, ions, amino acids, proteins and lipids in several tissue namely in brain and kidney through regulation of their transport and synthesis.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [1] | |
|
Johansson_61 Probe Info |
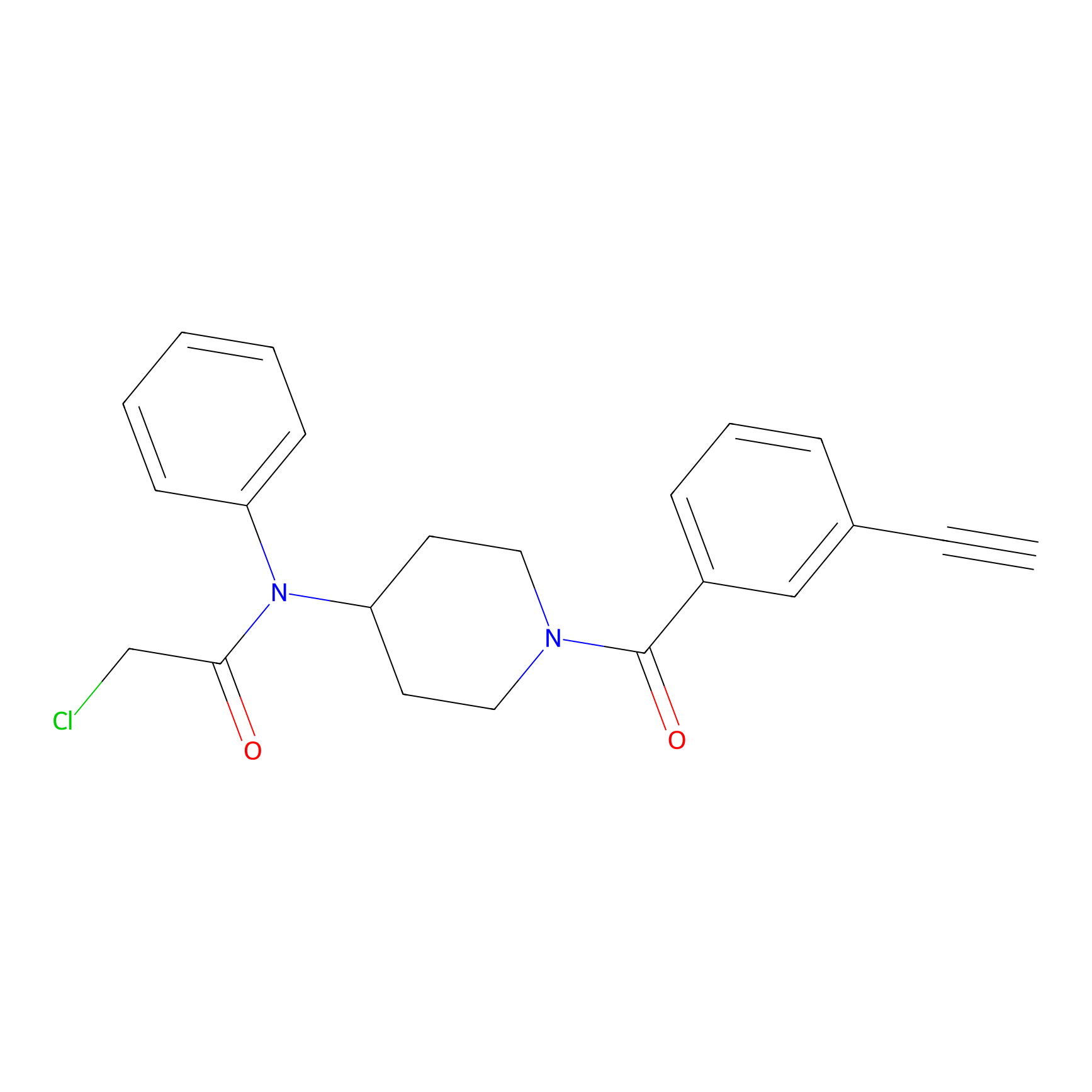 |
_(6.41) | LDD1485 | [2] | |
|
DBIA Probe Info |
 |
C92(1.11) | LDD1508 | [3] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C187 Probe Info |
 |
24.59 | LDD1865 | [4] | |
|
C191 Probe Info |
 |
18.64 | LDD1868 | [4] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0215 | AC10 | HEK-293T | C92(1.11) | LDD1508 | [3] |
| LDCM0270 | AC15 | HEK-293T | C92(1.17) | LDD1513 | [3] |
| LDCM0277 | AC18 | HEK-293T | C92(1.12) | LDD1516 | [3] |
| LDCM0279 | AC2 | HEK-293T | C92(0.99) | LDD1518 | [3] |
| LDCM0283 | AC23 | HEK-293T | C92(1.13) | LDD1522 | [3] |
| LDCM0286 | AC26 | HEK-293T | C92(1.09) | LDD1525 | [3] |
| LDCM0292 | AC31 | HEK-293T | C92(1.02) | LDD1531 | [3] |
| LDCM0295 | AC34 | HEK-293T | C92(1.04) | LDD1534 | [3] |
| LDCM0300 | AC39 | HEK-293T | C92(1.08) | LDD1539 | [3] |
| LDCM0304 | AC42 | HEK-293T | C92(1.02) | LDD1543 | [3] |
| LDCM0309 | AC47 | HEK-293T | C92(1.05) | LDD1548 | [3] |
| LDCM0313 | AC50 | HEK-293T | C92(1.07) | LDD1552 | [3] |
| LDCM0318 | AC55 | HEK-293T | C92(1.00) | LDD1557 | [3] |
| LDCM0321 | AC58 | HEK-293T | C92(1.13) | LDD1560 | [3] |
| LDCM0327 | AC63 | HEK-293T | C92(1.15) | LDD1566 | [3] |
| LDCM0334 | AC7 | HEK-293T | C92(1.13) | LDD1568 | [3] |
| LDCM0379 | CL11 | HEK-293T | C92(1.34) | LDD1583 | [3] |
| LDCM0405 | CL18 | HEK-293T | C92(1.09) | LDD1609 | [3] |
| LDCM0411 | CL23 | HEK-293T | C92(1.03) | LDD1615 | [3] |
| LDCM0419 | CL30 | HEK-293T | C92(1.03) | LDD1623 | [3] |
| LDCM0424 | CL35 | HEK-293T | C92(1.07) | LDD1628 | [3] |
| LDCM0432 | CL42 | HEK-293T | C92(0.93) | LDD1636 | [3] |
| LDCM0437 | CL47 | HEK-293T | C92(1.02) | LDD1641 | [3] |
| LDCM0445 | CL54 | HEK-293T | C92(2.00) | LDD1648 | [3] |
| LDCM0450 | CL59 | HEK-293T | C92(1.20) | LDD1653 | [3] |
| LDCM0451 | CL6 | HEK-293T | C92(1.65) | LDD1654 | [3] |
| LDCM0458 | CL66 | HEK-293T | C92(1.27) | LDD1661 | [3] |
| LDCM0464 | CL71 | HEK-293T | C92(1.04) | LDD1667 | [3] |
| LDCM0471 | CL78 | HEK-293T | C92(1.07) | LDD1674 | [3] |
| LDCM0477 | CL83 | HEK-293T | C92(1.29) | LDD1680 | [3] |
| LDCM0485 | CL90 | HEK-293T | C92(3.21) | LDD1688 | [3] |
| LDCM0490 | CL95 | HEK-293T | C92(2.46) | LDD1693 | [3] |
| LDCM0569 | Fragment7 | Jurkat | _(6.41) | LDD1485 | [2] |
The Interaction Atlas With This Target
References
