Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
TG42 Probe Info |
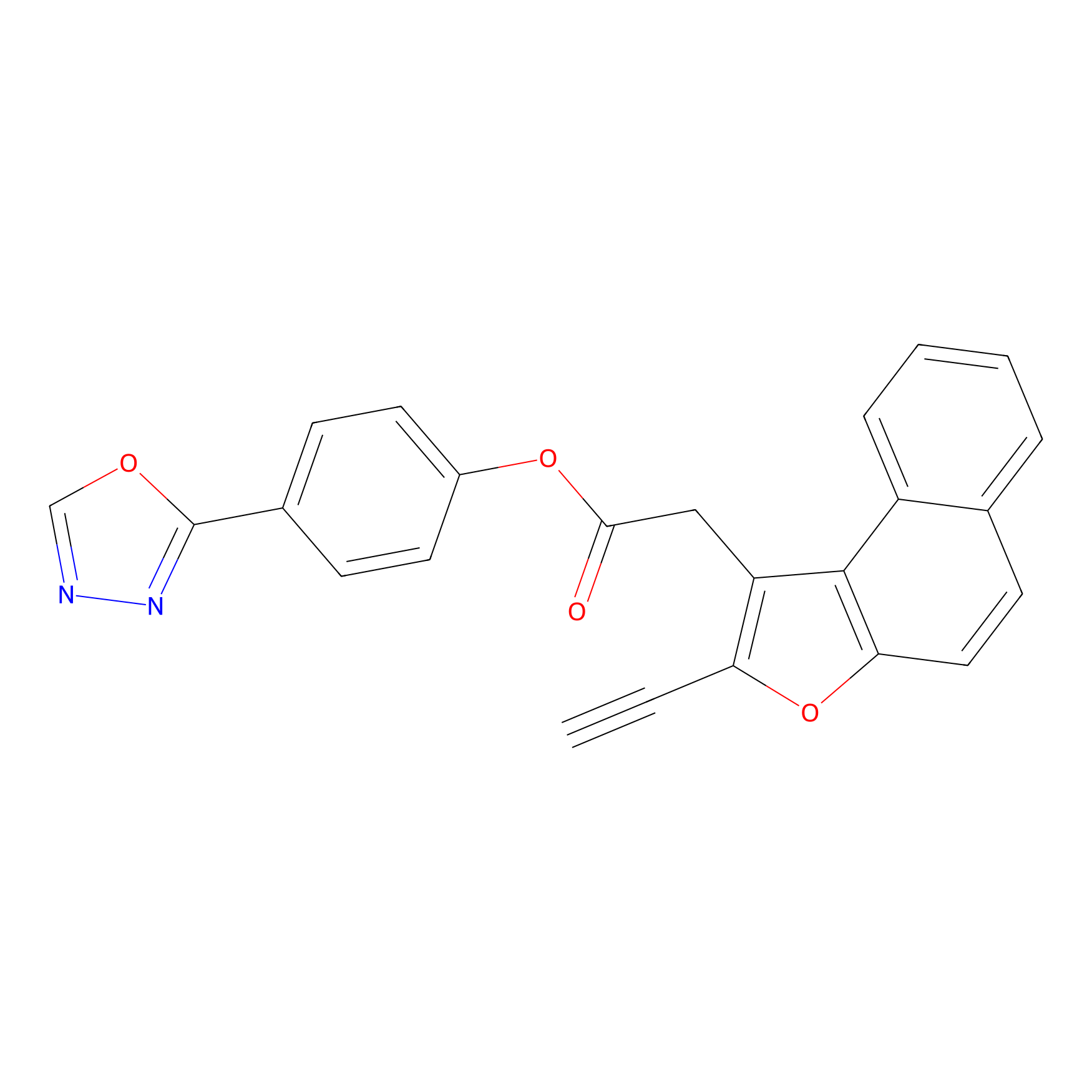 |
4.40 | LDD0326 | [1] | |
|
C-Sul Probe Info |
 |
19.33 | LDD0066 | [2] | |
|
AZ-9 Probe Info |
 |
E7(10.00) | LDD2209 | [3] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [4] | |
|
DBIA Probe Info |
 |
C20(1.01) | LDD3310 | [5] | |
|
BTD Probe Info |
 |
C20(0.94) | LDD2112 | [6] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [7] | |
|
ATP probe Probe Info |
 |
K17(0.00); K31(0.00); K92(0.00) | LDD0199 | [8] | |
|
m-APA Probe Info |
 |
N.A. | LDD2233 | [7] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [9] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [9] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [10] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [9] | |
|
JW-RF-010 Probe Info |
 |
N.A. | LDD0026 | [11] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [11] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [12] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [12] | |
|
NHS Probe Info |
 |
K92(0.00); K135(0.00) | LDD0010 | [12] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [13] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [12] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [14] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [15] | |
|
MPP-AC Probe Info |
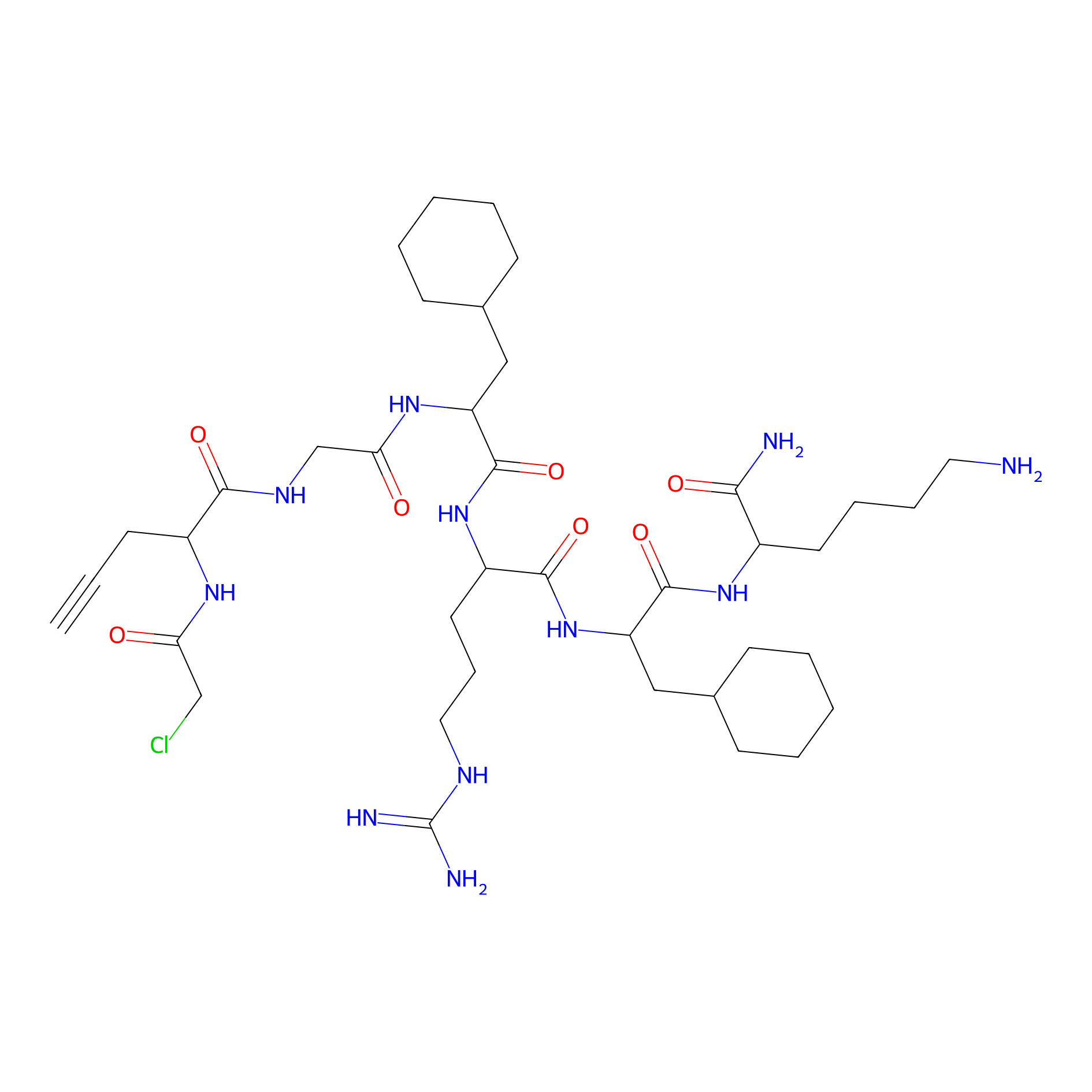 |
N.A. | LDD0428 | [16] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [17] | |
|
TER-AC Probe Info |
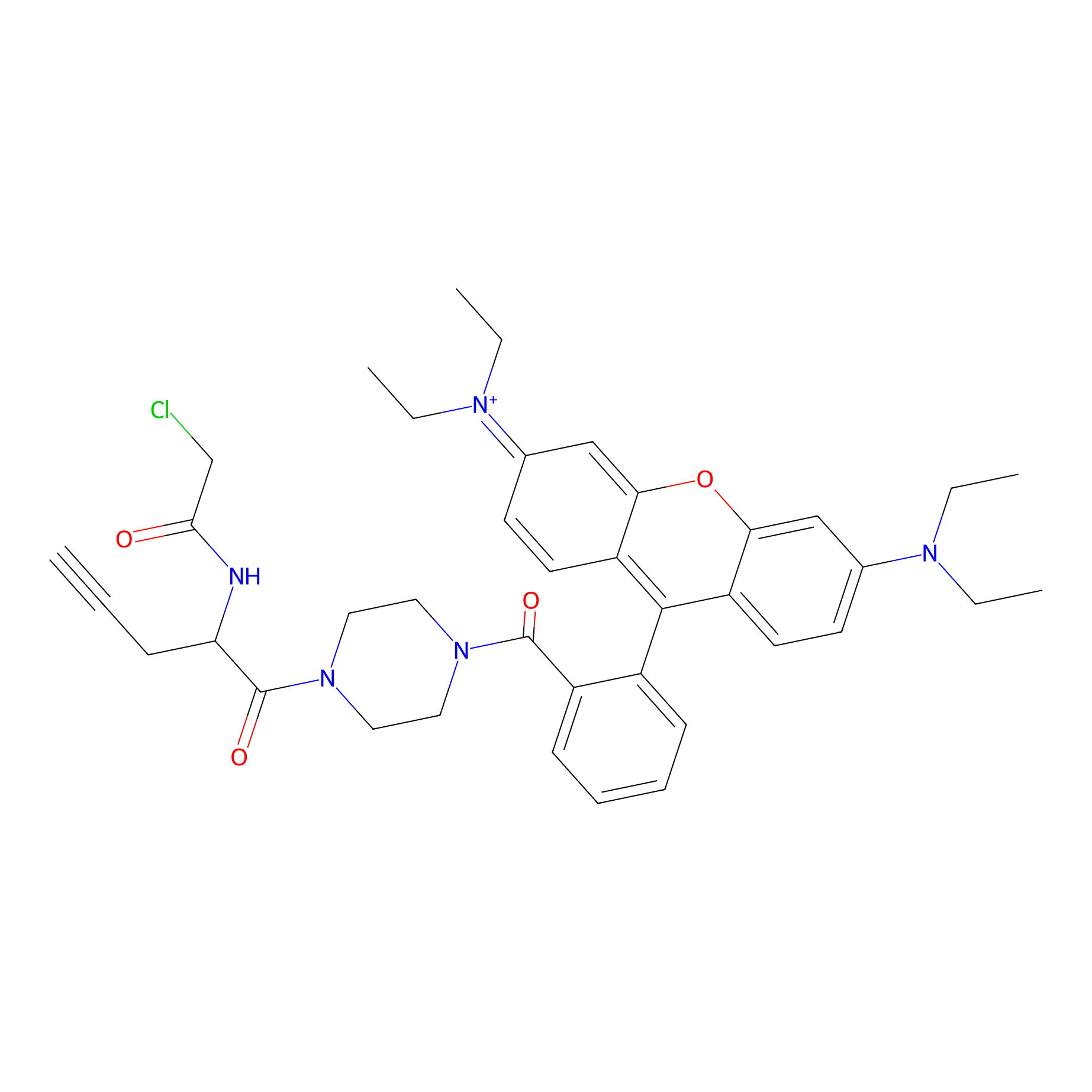 |
N.A. | LDD0426 | [16] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [18] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C20(0.94) | LDD2112 | [6] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C20(1.28) | LDD2117 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | H45(0.00); C20(0.00) | LDD0222 | [15] |
| LDCM0632 | CL-Sc | Hep-G2 | C20(0.84) | LDD2227 | [17] |
| LDCM0625 | F8 | Ramos | C20(1.76) | LDD2187 | [19] |
| LDCM0572 | Fragment10 | Ramos | C20(0.85) | LDD2189 | [19] |
| LDCM0573 | Fragment11 | Ramos | C20(0.40) | LDD2190 | [19] |
| LDCM0574 | Fragment12 | Ramos | C20(0.84) | LDD2191 | [19] |
| LDCM0575 | Fragment13 | Ramos | C20(1.61) | LDD2192 | [19] |
| LDCM0576 | Fragment14 | Ramos | C20(0.79) | LDD2193 | [19] |
| LDCM0579 | Fragment20 | Ramos | C20(0.64) | LDD2194 | [19] |
| LDCM0580 | Fragment21 | Ramos | C20(1.42) | LDD2195 | [19] |
| LDCM0582 | Fragment23 | Ramos | C20(1.92) | LDD2196 | [19] |
| LDCM0578 | Fragment27 | Ramos | C20(1.35) | LDD2197 | [19] |
| LDCM0586 | Fragment28 | Ramos | C20(0.46) | LDD2198 | [19] |
| LDCM0588 | Fragment30 | Ramos | C20(1.97) | LDD2199 | [19] |
| LDCM0589 | Fragment31 | Ramos | C20(1.19) | LDD2200 | [19] |
| LDCM0590 | Fragment32 | Ramos | C20(0.81) | LDD2201 | [19] |
| LDCM0468 | Fragment33 | Ramos | C20(1.39) | LDD2202 | [19] |
| LDCM0596 | Fragment38 | Ramos | C20(1.00) | LDD2203 | [19] |
| LDCM0566 | Fragment4 | Ramos | C20(0.66) | LDD2184 | [19] |
| LDCM0610 | Fragment52 | Ramos | C20(1.74) | LDD2204 | [19] |
| LDCM0614 | Fragment56 | Ramos | C20(2.27) | LDD2205 | [19] |
| LDCM0569 | Fragment7 | Ramos | C20(0.58) | LDD2186 | [19] |
| LDCM0571 | Fragment9 | Ramos | C20(0.75) | LDD2188 | [19] |
| LDCM0107 | IAA | HeLa | C20(0.00); H45(0.00) | LDD0221 | [15] |
| LDCM0022 | KB02 | Ramos | C20(0.63) | LDD2182 | [19] |
| LDCM0023 | KB03 | Ramos | C20(0.66) | LDD2183 | [19] |
| LDCM0024 | KB05 | COLO792 | C20(1.01) | LDD3310 | [5] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C20(0.60) | LDD2114 | [6] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C20(1.43) | LDD2125 | [6] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C20(0.62) | LDD2133 | [6] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C20(0.66) | LDD2136 | [6] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C20(0.56) | LDD2148 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| ATP-dependent RNA helicase DDX39A (DDX39A) | DEAD box helicase family | O00148 | |||
| Spliceosome RNA helicase DDX39B (DDX39B) | DEAD box helicase family | Q13838 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Wolframin (WFS1) | . | O76024 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Splicing factor U2AF 35 kDa subunit (U2AF1) | Splicing factor SR family | Q01081 | |||
| Splicing factor U2AF 65 kDa subunit (U2AF2) | Splicing factor SR family | P26368 | |||
References
