Details of the Target
General Information of Target
| Target ID | LDTP04084 | |||||
|---|---|---|---|---|---|---|
| Target Name | Probable 28S rRNA (cytosine(4447)-C(5))-methyltransferase (NOP2) | |||||
| Gene Name | NOP2 | |||||
| Gene ID | 4839 | |||||
| Synonyms |
NOL1; NSUN1; Probable 28S rRNA; cytosine(4447)-C(5))-methyltransferase; EC 2.1.1.-; Nucleolar protein 1; Nucleolar protein 2 homolog; Proliferating-cell nucleolar antigen p120; Proliferation-associated nucleolar protein p120
|
|||||
| 3D Structure | ||||||
| Sequence |
MGRKLDPTKEKRGPGRKARKQKGAETELVRFLPAVSDENSKRLSSRARKRAAKRRLGSVE
APKTNKSPEAKPLPGKLPKGISAGAVQTAGKKGPQSLFNAPRGKKRPAPGSDEEEEEEDS EEDGMVNHGDLWGSEDDADTVDDYGADSNSEDEEEGEALLPIERAARKQKAREAAAGIQW SEEETEDEEEEKEVTPESGPPKVEEADGGLQINVDEEPFVLPPAGEMEQDAQAPDLQRVH KRIQDIVGILRDFGAQREEGRSRSEYLNRLKKDLAIYYSYGDFLLGKLMDLFPLSELVEF LEANEVPRPVTLRTNTLKTRRRDLAQALINRGVNLDPLGKWSKTGLVVYDSSVPIGATPE YLAGHYMLQGASSMLPVMALAPQEHERILDMCCAPGGKTSYMAQLMKNTGVILANDANAE RLKSVVGNLHRLGVTNTIISHYDGRQFPKVVGGFDRVLLDAPCSGTGVISKDPAVKTNKD EKDILRCAHLQKELLLSAIDSVNATSKTGGYLVYCTCSITVEENEWVVDYALKKRNVRLV PTGLDFGQEGFTRFRERRFHPSLRSTRRFYPHTHNMDGFFIAKFKKFSNSIPQSQTGNSE TATPTNVDLPQVIPKSENSSQPAKKAKGAAKTKQQLQKQQHPKKASFQKLNGISKGADSE LSTVPSVTKTQASSSFQDSSQPAGKAEGIREPKVTGKLKQRSPKLQSSKKVAFLRQNAPP KGTDTQTPAVLSPSKTQATLKPKDHHQPLGRAKGVEKQQLPEQPFEKAAFQKQNDTPKGP QPPTVSPIRSSRPPPAKRKKSQSRGNSQLLLS |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Class I-like SAM-binding methyltransferase superfamily, RsmB/NOP family
|
|||||
| Subcellular location |
Nucleus, nucleolus
|
|||||
| Function |
Involved in ribosomal large subunit assembly. S-adenosyl-L-methionine-dependent methyltransferase that specifically methylates the C(5) position of cytosine 4447 in 28S rRNA (Probable). May play a role in the regulation of the cell cycle and the increased nucleolar activity that is associated with the cell proliferation (Probable).
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| C32 | Substitution: p.L131F | . | |||
| HCT15 | SNV: p.S497I | DBIA Probe Info | |||
| HEC1B | SNV: p.E296D | . | |||
| Ishikawa (Heraklio) 02 ER | SNV: p.E522K | DBIA Probe Info | |||
| JURKAT | SNV: p.L285F | Compound 10 Probe Info | |||
| KMCH1 | SNV: p.P359L | DBIA Probe Info | |||
| KNS81FD | SNV: p.D775Y | DBIA Probe Info | |||
| KYSE410 | SNV: p.S732F | DBIA Probe Info | |||
| NCIH1703 | Deletion: p.N618QfsTer61 | DBIA Probe Info | |||
| OVK18 | SNV: p.E259D | DBIA Probe Info | |||
| SKMEL28 | SNV: p.G286C | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
AZ-9 Probe Info |
 |
3.97 | LDD0393 | [1] | |
|
DBIA Probe Info |
 |
C520(3.34); C496(5.64) | LDD3311 | [2] | |
|
BTD Probe Info |
 |
C487(1.16) | LDD1700 | [3] | |
|
ATP probe Probe Info |
 |
K471(0.00); K63(0.00); K66(0.00) | LDD0199 | [4] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0358 | [5] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0032 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
C463(0.00); C392(0.00); C393(0.00) | LDD0037 | [6] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [8] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [9] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [9] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [10] | |
|
NHS Probe Info |
 |
K91(0.00); K767(0.00); K757(0.00); K669(0.00) | LDD0010 | [10] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [10] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [10] | |
|
1c-yne Probe Info |
 |
K757(0.00); K669(0.00); K471(0.00) | LDD0228 | [5] | |
|
Acrolein Probe Info |
 |
C463(0.00); C392(0.00); C393(0.00) | LDD0217 | [11] | |
|
Crotonaldehyde Probe Info |
 |
H745(0.00); C463(0.00) | LDD0219 | [11] | |
|
Methacrolein Probe Info |
 |
C463(0.00); C393(0.00) | LDD0218 | [11] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [12] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [14] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [14] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [15] | |
|
HM-2 Probe Info |
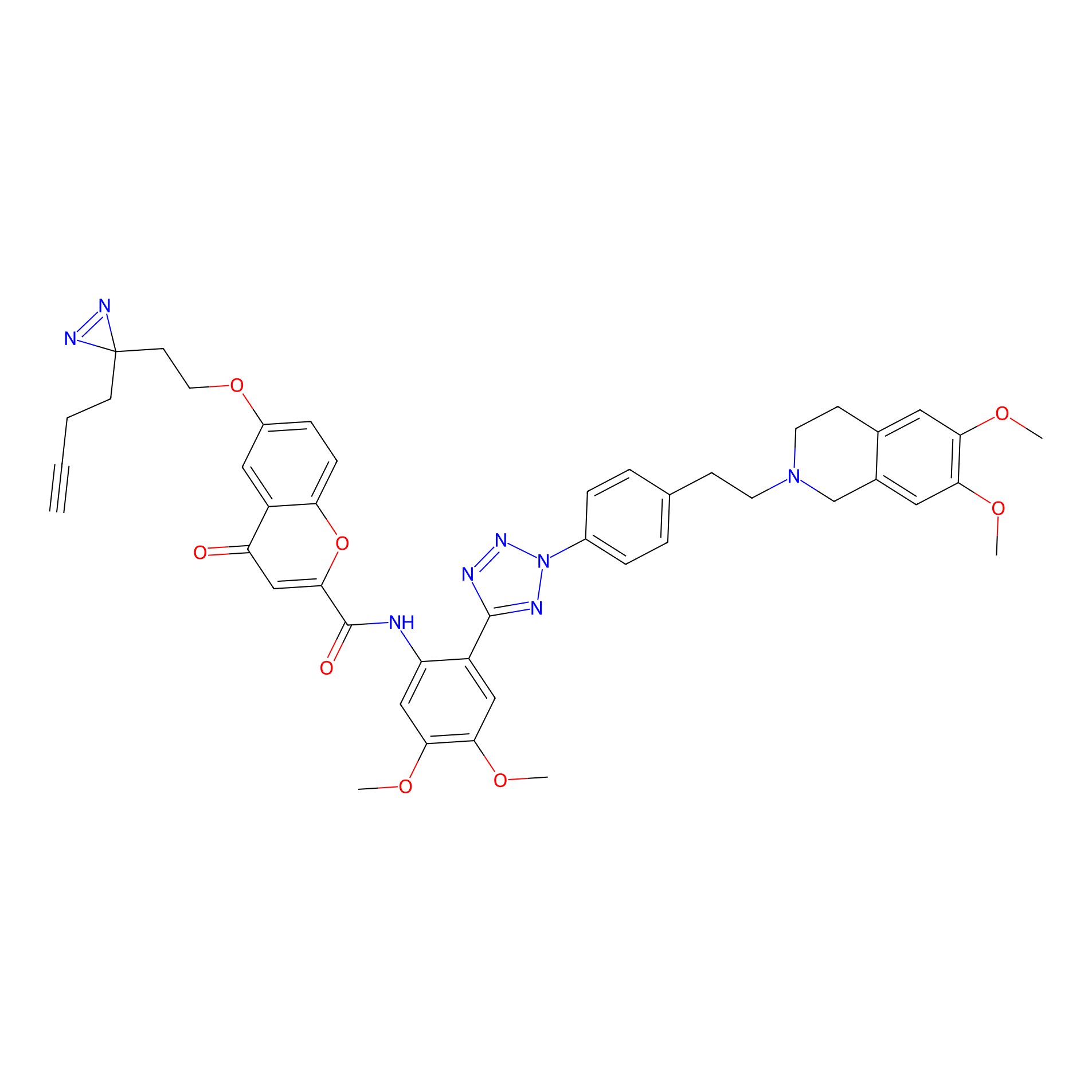 |
3.38 | LDD0436 | [16] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C487(0.57) | LDD2142 | [3] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C393(1.05) | LDD2112 | [3] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C463(0.77) | LDD2117 | [3] |
| LDCM0539 | 3-(4-Isopropylpiperazin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C487(0.56) | LDD2132 | [3] |
| LDCM0545 | Acetamide | MDA-MB-231 | C487(0.68) | LDD2138 | [3] |
| LDCM0108 | Chloroacetamide | HeLa | C463(0.00); C392(0.00); H441(0.00); H745(0.00) | LDD0222 | [11] |
| LDCM0632 | CL-Sc | Hep-G2 | C463(2.00) | LDD2227 | [13] |
| LDCM0625 | F8 | Ramos | C463(1.62) | LDD2187 | [17] |
| LDCM0573 | Fragment11 | Ramos | C463(0.48) | LDD2190 | [17] |
| LDCM0576 | Fragment14 | Ramos | C463(0.90) | LDD2193 | [17] |
| LDCM0566 | Fragment4 | Ramos | C463(1.24) | LDD2184 | [17] |
| LDCM0569 | Fragment7 | Ramos | C463(0.64) | LDD2186 | [17] |
| LDCM0173 | HM30181 | Hep-G2 | 3.38 | LDD0436 | [16] |
| LDCM0107 | IAA | HeLa | C392(0.00); C463(0.00); H745(0.00) | LDD0221 | [11] |
| LDCM0022 | KB02 | HEK-293T | C463(0.94) | LDD1492 | [18] |
| LDCM0023 | KB03 | HEK-293T | C463(1.01) | LDD1497 | [18] |
| LDCM0024 | KB05 | G361 | C520(3.34); C496(5.64) | LDD3311 | [2] |
| LDCM0109 | NEM | HeLa | H441(0.00); H745(0.00); H746(0.00) | LDD0223 | [11] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C463(0.79) | LDD2089 | [3] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C487(0.82); C463(0.85) | LDD2099 | [3] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C487(0.77) | LDD2100 | [3] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C487(0.56); C393(0.70) | LDD2104 | [3] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C487(0.35); C392(0.55); C393(0.69) | LDD2106 | [3] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C487(1.04); C463(1.07) | LDD2107 | [3] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C487(0.69) | LDD2108 | [3] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C487(0.44); C463(0.54); C393(0.95) | LDD2109 | [3] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C487(0.50) | LDD2114 | [3] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C487(0.72); C463(0.76) | LDD2120 | [3] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C487(0.71) | LDD2122 | [3] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C487(0.46); C463(0.91) | LDD2123 | [3] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C463(0.70); C393(1.06) | LDD2125 | [3] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C393(1.06) | LDD2127 | [3] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C487(0.83) | LDD2129 | [3] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C463(1.56) | LDD2135 | [3] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C463(1.07) | LDD2136 | [3] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C487(0.54); C463(1.10) | LDD2137 | [3] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C487(1.16) | LDD1700 | [3] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C487(0.62); C463(0.77) | LDD2140 | [3] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C487(0.96) | LDD2143 | [3] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C463(3.11) | LDD2144 | [3] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C487(0.96); C463(0.97) | LDD2146 | [3] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C392(0.98) | LDD2148 | [3] |
| LDCM0557 | Nucleophilic fragment 8b | MDA-MB-231 | C487(1.36) | LDD2151 | [3] |
| LDCM0113 | W17 | Hep-G2 | C463(0.60) | LDD0240 | [12] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Importin subunit alpha-7 (KPNA6) | Importin alpha family | O60684 | |||
Other
References
