Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [2] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [2] | |
|
W1 Probe Info |
 |
16.85 | LDD0235 | [3] | |
|
C-Sul Probe Info |
 |
2.47 | LDD0066 | [4] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [5] | |
|
STPyne Probe Info |
 |
K137(9.49); K143(4.42); K177(10.00); K196(6.25) | LDD0277 | [6] | |
|
AZ-9 Probe Info |
 |
E210(10.00) | LDD2208 | [7] | |
|
Sulforaphane-probe2 Probe Info |
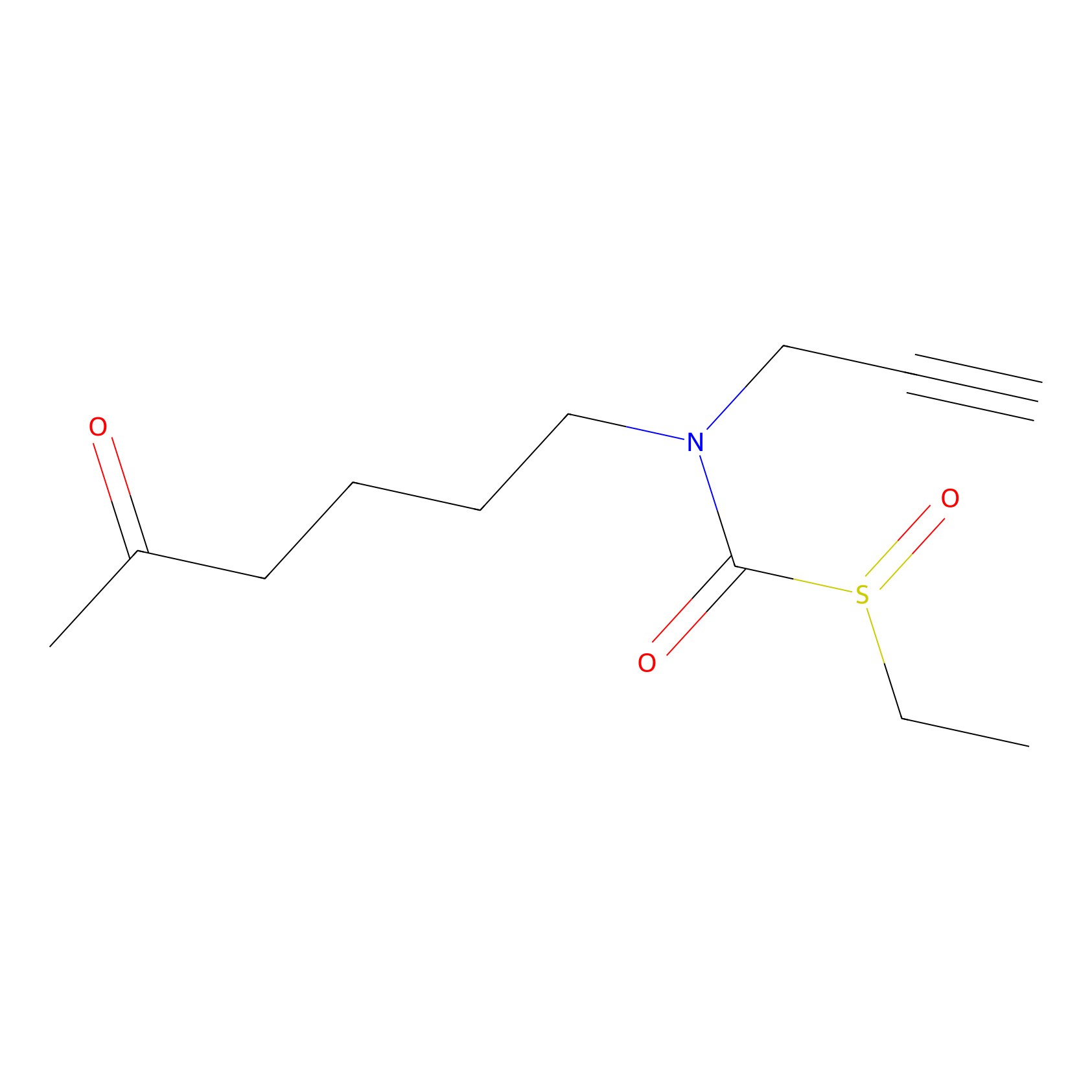 |
1.71 | LDD0160 | [8] | |
|
HHS-475 Probe Info |
 |
Y98(0.42) | LDD0264 | [9] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0223 | [10] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [11] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0151 | [12] | |
|
AOyne Probe Info |
 |
13.40 | LDD0443 | [13] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
20.00 | LDD0472 | [15] | |
|
DFG-out-3 Probe Info |
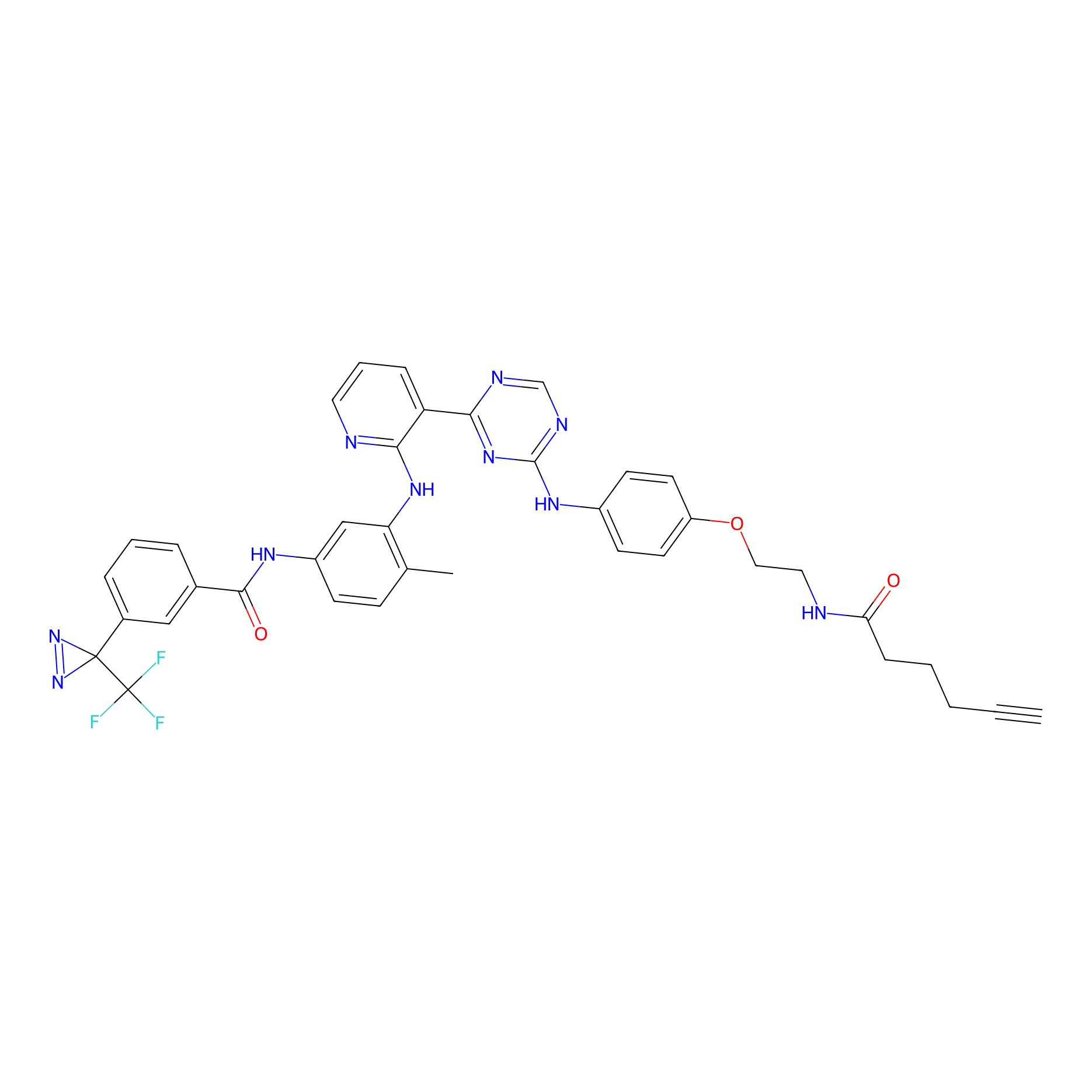 |
2.50 | LDD0074 | [16] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 14.07 | LDD0403 | [1] |
| LDCM0116 | HHS-0101 | DM93 | Y98(0.42) | LDD0264 | [9] |
| LDCM0117 | HHS-0201 | DM93 | Y98(0.41) | LDD0265 | [9] |
| LDCM0118 | HHS-0301 | DM93 | Y98(0.44) | LDD0266 | [9] |
| LDCM0119 | HHS-0401 | DM93 | Y98(0.52) | LDD0267 | [9] |
| LDCM0120 | HHS-0701 | DM93 | Y98(0.42) | LDD0268 | [9] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [10] |
| LDCM0016 | Ranjitkar_cp1 | A431 | 2.50 | LDD0074 | [16] |
| LDCM0003 | Sulforaphane | MDA-MB-231 | 1.71 | LDD0160 | [8] |
References
