Details of the Target
General Information of Target
| Target ID | LDTP03552 | |||||
|---|---|---|---|---|---|---|
| Target Name | Pyruvate kinase PKLR (PKLR) | |||||
| Gene Name | PKLR | |||||
| Gene ID | 5313 | |||||
| Synonyms |
PK1; PKL; Pyruvate kinase PKLR; EC 2.7.1.40; Pyruvate kinase 1; Pyruvate kinase isozymes L/R; R-type/L-type pyruvate kinase; Red cell/liver pyruvate kinase |
|||||
| 3D Structure | ||||||
| Sequence |
MSIQENISSLQLRSWVSKSQRDLAKSILIGAPGGPAGYLRRASVAQLTQELGTAFFQQQQ
LPAAMADTFLEHLCLLDIDSEPVAARSTSIIATIGPASRSVERLKEMIKAGMNIARLNFS HGSHEYHAESIANVREAVESFAGSPLSYRPVAIALDTKGPEIRTGILQGGPESEVELVKG SQVLVTVDPAFRTRGNANTVWVDYPNIVRVVPVGGRIYIDDGLISLVVQKIGPEGLVTQV ENGGVLGSRKGVNLPGAQVDLPGLSEQDVRDLRFGVEHGVDIVFASFVRKASDVAAVRAA LGPEGHGIKIISKIENHEGVKRFDEILEVSDGIMVARGDLGIEIPAEKVFLAQKMMIGRC NLAGKPVVCATQMLESMITKPRPTRAETSDVANAVLDGADCIMLSGETAKGNFPVEAVKM QHAIAREAEAAVYHRQLFEELRRAAPLSRDPTEVTAIGAVEAAFKCCAAAIIVLTTTGRS AQLLSRYRPRAAVIAVTRSAQAARQVHLCRGVFPLLYREPPEAIWADDVDRRVQFGIESG KLRGFLRVGDLVIVVTGWRPGSGYTNIMRVLSIS |
|||||
| Target Type |
Successful
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Pyruvate kinase family
|
|||||
| Function | Pyruvate kinase that catalyzes the conversion of phosphoenolpyruvate to pyruvate with the synthesis of ATP, and which plays a key role in glycolysis. | |||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
A-EBA Probe Info |
 |
3.40 | LDD0215 | [1] | |
|
STPyne Probe Info |
 |
K354(15.73) | LDD2217 | [2] | |
|
DBIA Probe Info |
 |
C509(1.24) | LDD3443 | [3] | |
|
HPAP Probe Info |
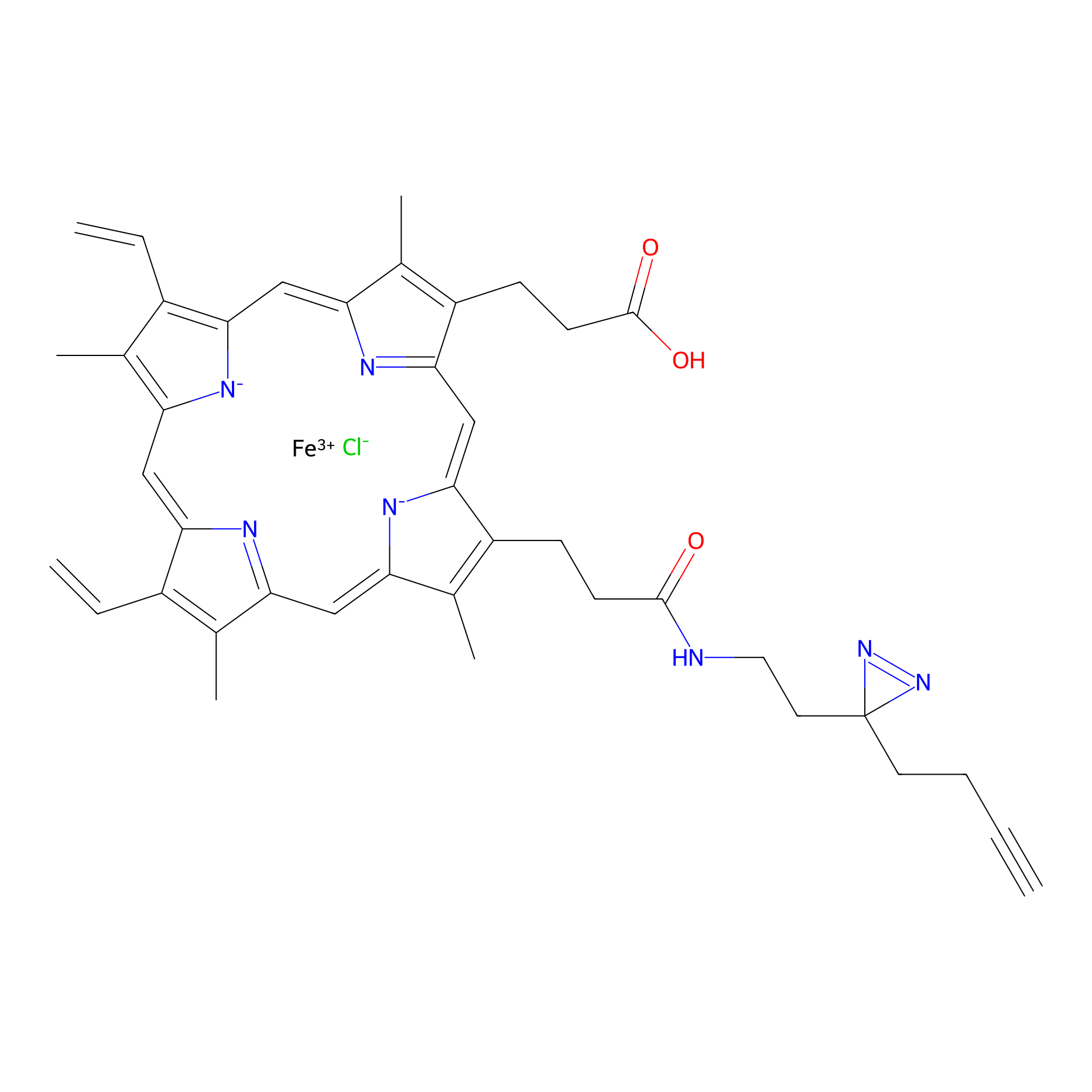 |
3.38 | LDD0063 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [6] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
8.41 | LDD0472 | [8] | |
|
FFF probe13 Probe Info |
 |
18.06 | LDD0476 | [8] | |
|
FFF probe3 Probe Info |
 |
12.23 | LDD0465 | [8] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [9] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Acyclovir | Small molecular drug | DB00787 | |||
| Pyruvic Acid | Small molecular drug | DB00119 | |||
| Mitapivat | . | DB16236 | |||
Investigative
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| 2-phosphoglycolic Acid | Small molecular drug | DB02726 | |||
| Beta-d-fructofuranose 16-bisphosphate | . | DB04551 | |||
References
