Details of the Target
General Information of Target
| Target ID | LDTP01153 | |||||
|---|---|---|---|---|---|---|
| Target Name | CCR4-NOT transcription complex subunit 3 (CNOT3) | |||||
| Gene Name | CNOT3 | |||||
| Gene ID | 4849 | |||||
| Synonyms |
KIAA0691; LENG2; NOT3; CCR4-NOT transcription complex subunit 3; CCR4-associated factor 3; Leukocyte receptor cluster member 2 |
|||||
| 3D Structure | ||||||
| Sequence |
MADKRKLQGEIDRCLKKVSEGVEQFEDIWQKLHNAANANQKEKYEADLKKEIKKLQRLRD
QIKTWVASNEIKDKRQLIDNRKLIETQMERFKVVERETKTKAYSKEGLGLAQKVDPAQKE KEEVGQWLTNTIDTLNMQVDQFESEVESLSVQTRKKKGDKDKQDRIEGLKRHIEKHRYHV RMLETILRMLDNDSILVDAIRKIKDDVEYYVDSSQDPDFEENEFLYDDLDLEDIPQALVA TSPPSHSHMEDEIFNQSSSTPTSTTSSSPIPPSPANCTTENSEDDKKRGRSTDSEVSQSP AKNGSKPVHSNQHPQSPAVPPTYPSGPPPAASALSTTPGNNGVPAPAAPPSALGPKASPA PSHNSGTPAPYAQAVAPPAPSGPSTTQPRPPSVQPSGGGGGGSGGGGSSSSSNSSAGGGA GKQNGATSYSSVVADSPAEVALSSSGGNNASSQALGPPSGPHNPPPSTSKEPSAAAPTGA GGVAPGSGNNSGGPSLLVPLPVNPPSSPTPSFSDAKAAGALLNGPPQFSTAPEIKAPEPL SSLKSMAERAAISSGIEDPVPTLHLTERDIILSSTSAPPASAQPPLQLSEVNIPLSLGVC PLGPVPLTKEQLYQQAMEEAAWHHMPHPSDSERIRQYLPRNPCPTPPYHHQMPPPHSDTV EFYQRLSTETLFFIFYYLEGTKAQYLAAKALKKQSWRFHTKYMMWFQRHEEPKTITDEFE QGTYIYFDYEKWGQRKKEGFTFEYRYLEDRDLQ |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
CNOT2/3/5 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Component of the CCR4-NOT complex which is one of the major cellular mRNA deadenylases and is linked to various cellular processes including bulk mRNA degradation, miRNA-mediated repression, translational repression during translational initiation and general transcription regulation. Additional complex functions may be a consequence of its influence on mRNA expression. May be involved in metabolic regulation; may be involved in recruitment of the CCR4-NOT complex to deadenylation target mRNAs involved in energy metabolism. Involved in mitotic progression and regulation of the spindle assembly checkpoint by regulating the stability of MAD1L1 mRNA. Can repress transcription and may link the CCR4-NOT complex to transcriptional regulation; the repressive function may involve histone deacetylases. Involved in the maintenance of embryonic stem (ES) cell identity.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K175(7.94); K41(5.51); K737(3.39); K92(5.78) | LDD0277 | [2] | |
|
DBIA Probe Info |
 |
C643(4.68) | LDD3373 | [3] | |
|
BTD Probe Info |
 |
C14(2.43) | LDD1700 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C600(4.05) | LDD0169 | [5] | |
|
IA-alkyne Probe Info |
 |
C600(7.78) | LDD1708 | [6] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [7] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [8] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [9] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [10] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C303 Probe Info |
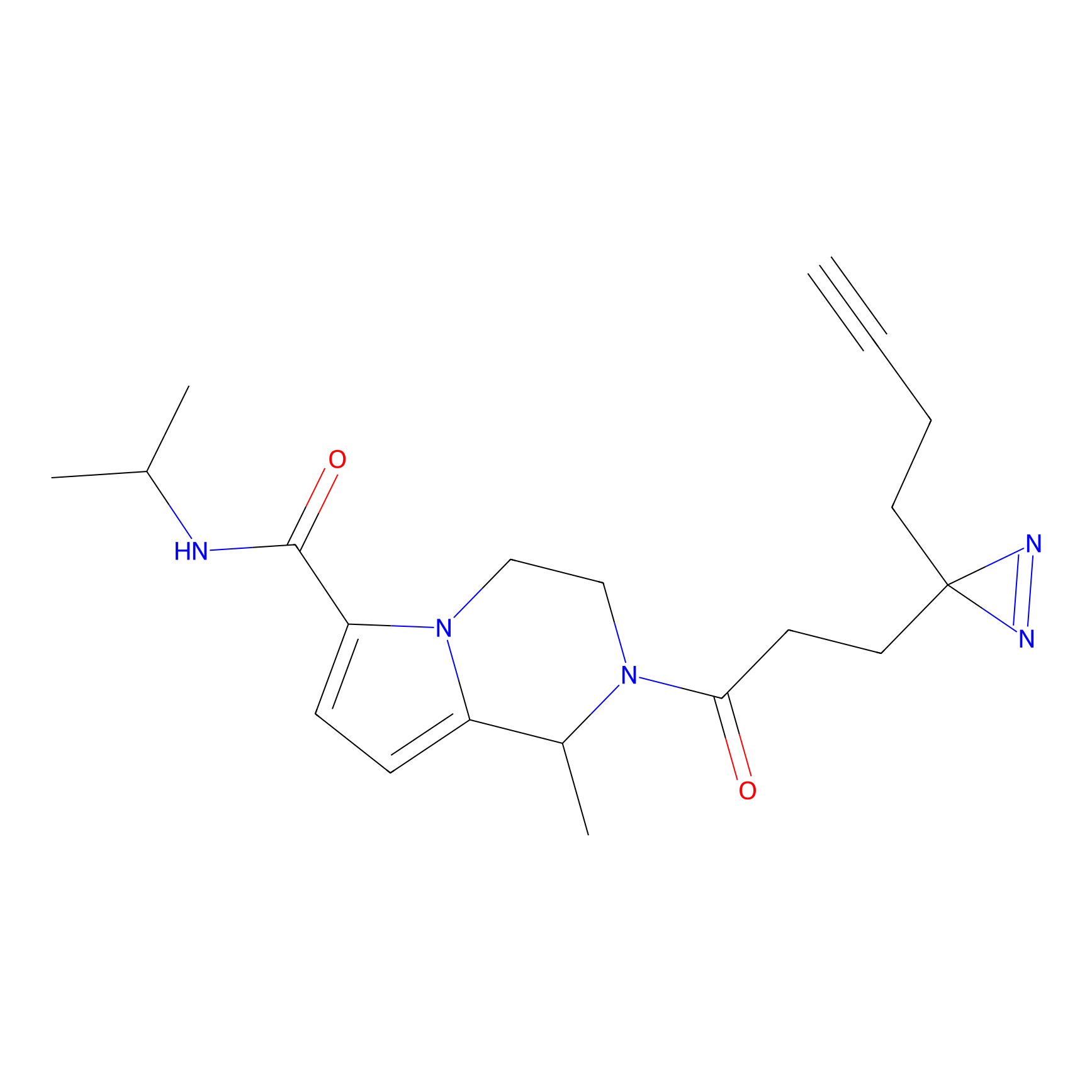 |
6.32 | LDD1972 | [12] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C14(1.05) | LDD2117 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C600(4.05) | LDD0169 | [5] |
| LDCM0214 | AC1 | HEK-293T | C600(1.09); C643(0.90) | LDD1507 | [13] |
| LDCM0215 | AC10 | HEK-293T | C600(1.16); C643(0.85) | LDD1508 | [13] |
| LDCM0226 | AC11 | HEK-293T | C600(0.99); C643(1.01) | LDD1509 | [13] |
| LDCM0237 | AC12 | HEK-293T | C600(0.95); C643(1.13) | LDD1510 | [13] |
| LDCM0259 | AC14 | HEK-293T | C600(1.08); C643(1.11) | LDD1512 | [13] |
| LDCM0276 | AC17 | HEK-293T | C600(1.03); C643(1.09) | LDD1515 | [13] |
| LDCM0277 | AC18 | HEK-293T | C600(1.15); C643(0.81) | LDD1516 | [13] |
| LDCM0278 | AC19 | HEK-293T | C600(1.20); C643(1.29) | LDD1517 | [13] |
| LDCM0279 | AC2 | HEK-293T | C600(0.99); C643(1.04) | LDD1518 | [13] |
| LDCM0280 | AC20 | HEK-293T | C600(1.25); C643(0.99) | LDD1519 | [13] |
| LDCM0281 | AC21 | HEK-293T | C600(0.85); C643(1.11) | LDD1520 | [13] |
| LDCM0282 | AC22 | HEK-293T | C600(1.12); C643(0.93) | LDD1521 | [13] |
| LDCM0284 | AC24 | HEK-293T | C643(1.04) | LDD1523 | [13] |
| LDCM0285 | AC25 | HEK-293T | C600(1.02); C643(1.03) | LDD1524 | [13] |
| LDCM0286 | AC26 | HEK-293T | C600(1.01); C643(0.92) | LDD1525 | [13] |
| LDCM0287 | AC27 | HEK-293T | C600(1.17); C643(1.26) | LDD1526 | [13] |
| LDCM0288 | AC28 | HEK-293T | C600(0.89); C643(1.06) | LDD1527 | [13] |
| LDCM0289 | AC29 | HEK-293T | C600(0.88); C643(1.07) | LDD1528 | [13] |
| LDCM0290 | AC3 | HEK-293T | C600(1.06); C643(1.15) | LDD1529 | [13] |
| LDCM0291 | AC30 | HEK-293T | C600(1.03); C643(0.96) | LDD1530 | [13] |
| LDCM0293 | AC32 | HEK-293T | C643(0.95) | LDD1532 | [13] |
| LDCM0294 | AC33 | HEK-293T | C600(1.11); C643(1.16) | LDD1533 | [13] |
| LDCM0295 | AC34 | HEK-293T | C600(0.94); C643(0.92) | LDD1534 | [13] |
| LDCM0296 | AC35 | HEK-293T | C600(1.13); C643(1.00) | LDD1535 | [13] |
| LDCM0297 | AC36 | HEK-293T | C600(1.04); C643(1.02) | LDD1536 | [13] |
| LDCM0298 | AC37 | HEK-293T | C600(0.87); C643(0.98) | LDD1537 | [13] |
| LDCM0299 | AC38 | HEK-293T | C600(0.99); C643(1.07) | LDD1538 | [13] |
| LDCM0301 | AC4 | HEK-293T | C600(0.98); C643(1.12) | LDD1540 | [13] |
| LDCM0302 | AC40 | HEK-293T | C643(0.97) | LDD1541 | [13] |
| LDCM0303 | AC41 | HEK-293T | C600(1.06); C643(1.06) | LDD1542 | [13] |
| LDCM0304 | AC42 | HEK-293T | C600(1.04); C643(0.96) | LDD1543 | [13] |
| LDCM0305 | AC43 | HEK-293T | C600(0.97); C643(0.99) | LDD1544 | [13] |
| LDCM0306 | AC44 | HEK-293T | C600(1.07); C643(1.12) | LDD1545 | [13] |
| LDCM0307 | AC45 | HEK-293T | C600(0.90); C643(0.96) | LDD1546 | [13] |
| LDCM0308 | AC46 | HEK-293T | C600(1.06); C643(1.12) | LDD1547 | [13] |
| LDCM0310 | AC48 | HEK-293T | C643(1.08) | LDD1549 | [13] |
| LDCM0311 | AC49 | HEK-293T | C600(1.12); C643(0.96) | LDD1550 | [13] |
| LDCM0312 | AC5 | HEK-293T | C600(0.96); C643(0.98) | LDD1551 | [13] |
| LDCM0313 | AC50 | HEK-293T | C600(1.10); C643(0.89) | LDD1552 | [13] |
| LDCM0314 | AC51 | HEK-293T | C600(1.16); C643(1.13) | LDD1553 | [13] |
| LDCM0315 | AC52 | HEK-293T | C600(1.04); C643(1.05) | LDD1554 | [13] |
| LDCM0316 | AC53 | HEK-293T | C600(0.89); C643(1.06) | LDD1555 | [13] |
| LDCM0317 | AC54 | HEK-293T | C600(1.02); C643(1.14) | LDD1556 | [13] |
| LDCM0319 | AC56 | HEK-293T | C643(0.96) | LDD1558 | [13] |
| LDCM0320 | AC57 | HEK-293T | C600(1.08); C643(1.05) | LDD1559 | [13] |
| LDCM0321 | AC58 | HEK-293T | C600(1.06); C643(0.92) | LDD1560 | [13] |
| LDCM0322 | AC59 | HEK-293T | C600(1.15); C643(1.04) | LDD1561 | [13] |
| LDCM0323 | AC6 | HEK-293T | C600(1.03); C643(1.03) | LDD1562 | [13] |
| LDCM0324 | AC60 | HEK-293T | C600(1.10); C643(1.01) | LDD1563 | [13] |
| LDCM0325 | AC61 | HEK-293T | C600(0.97); C643(1.10) | LDD1564 | [13] |
| LDCM0326 | AC62 | HEK-293T | C600(1.02); C643(1.14) | LDD1565 | [13] |
| LDCM0328 | AC64 | HEK-293T | C643(1.07) | LDD1567 | [13] |
| LDCM0345 | AC8 | HEK-293T | C643(1.02) | LDD1569 | [13] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C14(1.00) | LDD2113 | [4] |
| LDCM0248 | AKOS034007472 | HEK-293T | C600(0.98); C643(0.99) | LDD1511 | [13] |
| LDCM0356 | AKOS034007680 | HEK-293T | C600(1.15); C643(1.06) | LDD1570 | [13] |
| LDCM0275 | AKOS034007705 | HEK-293T | C643(0.94) | LDD1514 | [13] |
| LDCM0156 | Aniline | NCI-H1299 | 13.85 | LDD0403 | [1] |
| LDCM0367 | CL1 | HEK-293T | C600(1.20); C643(0.96) | LDD1571 | [13] |
| LDCM0368 | CL10 | HEK-293T | C600(1.04); C643(1.34) | LDD1572 | [13] |
| LDCM0369 | CL100 | HEK-293T | C600(1.02); C643(0.97) | LDD1573 | [13] |
| LDCM0370 | CL101 | HEK-293T | C600(1.16); C643(0.97) | LDD1574 | [13] |
| LDCM0371 | CL102 | HEK-293T | C600(1.13); C643(1.00) | LDD1575 | [13] |
| LDCM0372 | CL103 | HEK-293T | C600(1.03); C643(0.97) | LDD1576 | [13] |
| LDCM0373 | CL104 | HEK-293T | C600(0.92); C643(0.96) | LDD1577 | [13] |
| LDCM0374 | CL105 | HEK-293T | C600(1.14); C643(0.88) | LDD1578 | [13] |
| LDCM0375 | CL106 | HEK-293T | C600(0.99); C643(0.99) | LDD1579 | [13] |
| LDCM0376 | CL107 | HEK-293T | C600(0.98); C643(0.96) | LDD1580 | [13] |
| LDCM0377 | CL108 | HEK-293T | C600(1.00); C643(0.98) | LDD1581 | [13] |
| LDCM0378 | CL109 | HEK-293T | C600(1.15); C643(0.92) | LDD1582 | [13] |
| LDCM0380 | CL110 | HEK-293T | C600(1.30); C643(0.91) | LDD1584 | [13] |
| LDCM0381 | CL111 | HEK-293T | C600(1.12); C643(0.97) | LDD1585 | [13] |
| LDCM0382 | CL112 | HEK-293T | C600(1.09); C643(0.87) | LDD1586 | [13] |
| LDCM0383 | CL113 | HEK-293T | C600(1.18); C643(0.92) | LDD1587 | [13] |
| LDCM0384 | CL114 | HEK-293T | C600(1.13); C643(0.97) | LDD1588 | [13] |
| LDCM0385 | CL115 | HEK-293T | C600(1.10); C643(1.03) | LDD1589 | [13] |
| LDCM0386 | CL116 | HEK-293T | C600(0.91); C643(1.18) | LDD1590 | [13] |
| LDCM0387 | CL117 | HEK-293T | C600(1.25); C643(0.90) | LDD1591 | [13] |
| LDCM0388 | CL118 | HEK-293T | C600(1.18); C643(1.01) | LDD1592 | [13] |
| LDCM0389 | CL119 | HEK-293T | C600(1.00); C643(1.12) | LDD1593 | [13] |
| LDCM0390 | CL12 | HEK-293T | C643(1.04) | LDD1594 | [13] |
| LDCM0391 | CL120 | HEK-293T | C600(0.99); C643(1.13) | LDD1595 | [13] |
| LDCM0392 | CL121 | HEK-293T | C600(1.16); C643(1.03) | LDD1596 | [13] |
| LDCM0393 | CL122 | HEK-293T | C600(1.04); C643(1.10) | LDD1597 | [13] |
| LDCM0394 | CL123 | HEK-293T | C600(1.40); C643(0.90) | LDD1598 | [13] |
| LDCM0395 | CL124 | HEK-293T | C600(1.09); C643(1.00) | LDD1599 | [13] |
| LDCM0396 | CL125 | HEK-293T | C600(1.25); C643(0.98) | LDD1600 | [13] |
| LDCM0397 | CL126 | HEK-293T | C600(1.06); C643(1.01) | LDD1601 | [13] |
| LDCM0398 | CL127 | HEK-293T | C600(1.09); C643(0.96) | LDD1602 | [13] |
| LDCM0399 | CL128 | HEK-293T | C600(1.07); C643(0.98) | LDD1603 | [13] |
| LDCM0400 | CL13 | HEK-293T | C600(1.17); C643(1.07) | LDD1604 | [13] |
| LDCM0401 | CL14 | HEK-293T | C600(1.10); C643(1.05) | LDD1605 | [13] |
| LDCM0402 | CL15 | HEK-293T | C600(1.16); C643(1.19) | LDD1606 | [13] |
| LDCM0403 | CL16 | HEK-293T | C600(1.00); C643(0.96) | LDD1607 | [13] |
| LDCM0404 | CL17 | HEK-293T | C600(1.25); C643(1.24) | LDD1608 | [13] |
| LDCM0405 | CL18 | HEK-293T | C600(1.08); C643(0.97) | LDD1609 | [13] |
| LDCM0406 | CL19 | HEK-293T | C600(0.94); C643(1.07) | LDD1610 | [13] |
| LDCM0407 | CL2 | HEK-293T | C600(1.07); C643(1.12) | LDD1611 | [13] |
| LDCM0408 | CL20 | HEK-293T | C600(0.93); C643(0.95) | LDD1612 | [13] |
| LDCM0409 | CL21 | HEK-293T | C600(0.94); C643(0.78) | LDD1613 | [13] |
| LDCM0410 | CL22 | HEK-293T | C600(1.10); C643(1.24) | LDD1614 | [13] |
| LDCM0412 | CL24 | HEK-293T | C643(1.21) | LDD1616 | [13] |
| LDCM0413 | CL25 | HEK-293T | C600(1.28); C643(1.02) | LDD1617 | [13] |
| LDCM0414 | CL26 | HEK-293T | C600(1.08); C643(0.97) | LDD1618 | [13] |
| LDCM0415 | CL27 | HEK-293T | C600(1.13); C643(0.91) | LDD1619 | [13] |
| LDCM0416 | CL28 | HEK-293T | C600(0.97); C643(0.95) | LDD1620 | [13] |
| LDCM0417 | CL29 | HEK-293T | C600(0.93); C643(0.98) | LDD1621 | [13] |
| LDCM0418 | CL3 | HEK-293T | C600(1.04); C643(1.00) | LDD1622 | [13] |
| LDCM0419 | CL30 | HEK-293T | C600(1.00); C643(0.92) | LDD1623 | [13] |
| LDCM0420 | CL31 | HEK-293T | C600(1.12); C643(1.08) | LDD1624 | [13] |
| LDCM0421 | CL32 | HEK-293T | C600(0.81); C643(0.96) | LDD1625 | [13] |
| LDCM0422 | CL33 | HEK-293T | C600(1.23); C643(0.83) | LDD1626 | [13] |
| LDCM0423 | CL34 | HEK-293T | C600(0.91); C643(1.04) | LDD1627 | [13] |
| LDCM0425 | CL36 | HEK-293T | C643(1.04) | LDD1629 | [13] |
| LDCM0426 | CL37 | HEK-293T | C600(1.17); C643(0.91) | LDD1630 | [13] |
| LDCM0428 | CL39 | HEK-293T | C600(1.11); C643(0.97) | LDD1632 | [13] |
| LDCM0429 | CL4 | HEK-293T | C600(1.11); C643(1.04) | LDD1633 | [13] |
| LDCM0430 | CL40 | HEK-293T | C600(0.91); C643(1.01) | LDD1634 | [13] |
| LDCM0431 | CL41 | HEK-293T | C600(1.06); C643(1.00) | LDD1635 | [13] |
| LDCM0432 | CL42 | HEK-293T | C600(0.95); C643(0.89) | LDD1636 | [13] |
| LDCM0433 | CL43 | HEK-293T | C600(0.89); C643(0.99) | LDD1637 | [13] |
| LDCM0434 | CL44 | HEK-293T | C600(1.06); C643(0.92) | LDD1638 | [13] |
| LDCM0435 | CL45 | HEK-293T | C600(1.13); C643(0.84) | LDD1639 | [13] |
| LDCM0436 | CL46 | HEK-293T | C600(0.90); C643(1.14) | LDD1640 | [13] |
| LDCM0438 | CL48 | HEK-293T | C643(1.01) | LDD1642 | [13] |
| LDCM0439 | CL49 | HEK-293T | C600(0.99); C643(1.03) | LDD1643 | [13] |
| LDCM0440 | CL5 | HEK-293T | C600(1.04); C643(1.08) | LDD1644 | [13] |
| LDCM0441 | CL50 | HEK-293T | C600(1.11); C643(1.04) | LDD1645 | [13] |
| LDCM0443 | CL52 | HEK-293T | C600(0.92); C643(1.01) | LDD1646 | [13] |
| LDCM0444 | CL53 | HEK-293T | C600(1.04); C643(1.06) | LDD1647 | [13] |
| LDCM0445 | CL54 | HEK-293T | C600(1.25); C643(1.11) | LDD1648 | [13] |
| LDCM0446 | CL55 | HEK-293T | C600(0.98); C643(1.08) | LDD1649 | [13] |
| LDCM0447 | CL56 | HEK-293T | C600(1.05); C643(0.90) | LDD1650 | [13] |
| LDCM0448 | CL57 | HEK-293T | C600(1.00); C643(0.76) | LDD1651 | [13] |
| LDCM0449 | CL58 | HEK-293T | C600(0.90); C643(1.08) | LDD1652 | [13] |
| LDCM0451 | CL6 | HEK-293T | C600(1.13); C643(1.06) | LDD1654 | [13] |
| LDCM0452 | CL60 | HEK-293T | C643(1.00) | LDD1655 | [13] |
| LDCM0453 | CL61 | HEK-293T | C600(1.17); C643(0.93) | LDD1656 | [13] |
| LDCM0454 | CL62 | HEK-293T | C600(1.07); C643(1.01) | LDD1657 | [13] |
| LDCM0455 | CL63 | HEK-293T | C600(1.14); C643(0.92) | LDD1658 | [13] |
| LDCM0456 | CL64 | HEK-293T | C600(0.98); C643(0.96) | LDD1659 | [13] |
| LDCM0457 | CL65 | HEK-293T | C600(1.03); C643(0.90) | LDD1660 | [13] |
| LDCM0458 | CL66 | HEK-293T | C600(1.06); C643(0.95) | LDD1661 | [13] |
| LDCM0459 | CL67 | HEK-293T | C600(1.04); C643(1.18) | LDD1662 | [13] |
| LDCM0460 | CL68 | HEK-293T | C600(1.20); C643(0.99) | LDD1663 | [13] |
| LDCM0461 | CL69 | HEK-293T | C600(0.93); C643(0.86) | LDD1664 | [13] |
| LDCM0462 | CL7 | HEK-293T | C600(1.02); C643(1.11) | LDD1665 | [13] |
| LDCM0463 | CL70 | HEK-293T | C600(1.05); C643(1.06) | LDD1666 | [13] |
| LDCM0465 | CL72 | HEK-293T | C643(0.95) | LDD1668 | [13] |
| LDCM0466 | CL73 | HEK-293T | C600(1.28); C643(1.02) | LDD1669 | [13] |
| LDCM0467 | CL74 | HEK-293T | C600(1.13); C643(1.04) | LDD1670 | [13] |
| LDCM0469 | CL76 | HEK-293T | C600(0.93); C643(0.91) | LDD1672 | [13] |
| LDCM0470 | CL77 | HEK-293T | C600(1.14); C643(0.97) | LDD1673 | [13] |
| LDCM0471 | CL78 | HEK-293T | C600(1.03); C643(0.86) | LDD1674 | [13] |
| LDCM0472 | CL79 | HEK-293T | C600(0.97); C643(1.13) | LDD1675 | [13] |
| LDCM0473 | CL8 | HEK-293T | C600(1.25); C643(0.81) | LDD1676 | [13] |
| LDCM0474 | CL80 | HEK-293T | C600(1.21); C643(1.03) | LDD1677 | [13] |
| LDCM0475 | CL81 | HEK-293T | C600(0.81); C643(0.90) | LDD1678 | [13] |
| LDCM0476 | CL82 | HEK-293T | C600(0.97); C643(1.01) | LDD1679 | [13] |
| LDCM0478 | CL84 | HEK-293T | C643(0.98) | LDD1681 | [13] |
| LDCM0479 | CL85 | HEK-293T | C600(1.04); C643(0.99) | LDD1682 | [13] |
| LDCM0480 | CL86 | HEK-293T | C600(1.19); C643(0.95) | LDD1683 | [13] |
| LDCM0481 | CL87 | HEK-293T | C600(1.12); C643(1.06) | LDD1684 | [13] |
| LDCM0482 | CL88 | HEK-293T | C600(0.94); C643(0.95) | LDD1685 | [13] |
| LDCM0483 | CL89 | HEK-293T | C600(1.16); C643(1.03) | LDD1686 | [13] |
| LDCM0484 | CL9 | HEK-293T | C600(0.94); C643(0.86) | LDD1687 | [13] |
| LDCM0485 | CL90 | HEK-293T | C600(1.80); C643(1.61) | LDD1688 | [13] |
| LDCM0486 | CL91 | HEK-293T | C600(1.09); C643(1.15) | LDD1689 | [13] |
| LDCM0487 | CL92 | HEK-293T | C600(1.04); C643(0.93) | LDD1690 | [13] |
| LDCM0488 | CL93 | HEK-293T | C600(0.94); C643(0.97) | LDD1691 | [13] |
| LDCM0489 | CL94 | HEK-293T | C600(0.99); C643(0.93) | LDD1692 | [13] |
| LDCM0491 | CL96 | HEK-293T | C643(1.04) | LDD1694 | [13] |
| LDCM0492 | CL97 | HEK-293T | C600(1.07); C643(0.96) | LDD1695 | [13] |
| LDCM0493 | CL98 | HEK-293T | C600(1.11); C643(1.02) | LDD1696 | [13] |
| LDCM0494 | CL99 | HEK-293T | C600(1.19); C643(0.97) | LDD1697 | [13] |
| LDCM0495 | E2913 | HEK-293T | C600(1.47); C643(0.96) | LDD1698 | [13] |
| LDCM0468 | Fragment33 | HEK-293T | C600(1.07); C643(0.89) | LDD1671 | [13] |
| LDCM0427 | Fragment51 | HEK-293T | C600(1.03); C643(0.83) | LDD1631 | [13] |
| LDCM0022 | KB02 | HEK-293T | C600(0.86); C643(1.06) | LDD1492 | [13] |
| LDCM0023 | KB03 | HEK-293T | C600(1.00); C643(1.00) | LDD1497 | [13] |
| LDCM0024 | KB05 | OCUG-1 | C643(4.68) | LDD3373 | [3] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0225 | [10] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C14(0.91) | LDD2101 | [4] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C14(0.30) | LDD2104 | [4] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C14(0.34) | LDD2106 | [4] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C14(1.37) | LDD2107 | [4] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C14(0.32) | LDD2108 | [4] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C14(0.60) | LDD2109 | [4] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C14(1.34) | LDD2111 | [4] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C14(0.46) | LDD2115 | [4] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C14(3.22) | LDD2119 | [4] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C14(0.76) | LDD2123 | [4] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C14(0.83) | LDD2125 | [4] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C14(1.00) | LDD2129 | [4] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C14(0.51) | LDD2134 | [4] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C14(1.40) | LDD2135 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C14(1.06) | LDD2136 | [4] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C14(0.90) | LDD2137 | [4] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C14(2.43) | LDD1700 | [4] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C14(1.16) | LDD2140 | [4] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C14(4.53) | LDD2144 | [4] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C14(0.89) | LDD2146 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Calreticulin (CALR) | Calreticulin family | P27797 | |||
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Platelet endothelial cell adhesion molecule (PECAM1) | . | P16284 | |||
Other
References
