Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [2] | |
|
STPyne Probe Info |
 |
K174(7.69); K178(8.14); K188(7.58); K189(5.85) | LDD0277 | [3] | |
|
DBIA Probe Info |
 |
C148(1.50) | LDD3401 | [4] | |
|
BTD Probe Info |
 |
C296(1.48) | LDD2118 | [5] | |
|
Johansson_61 Probe Info |
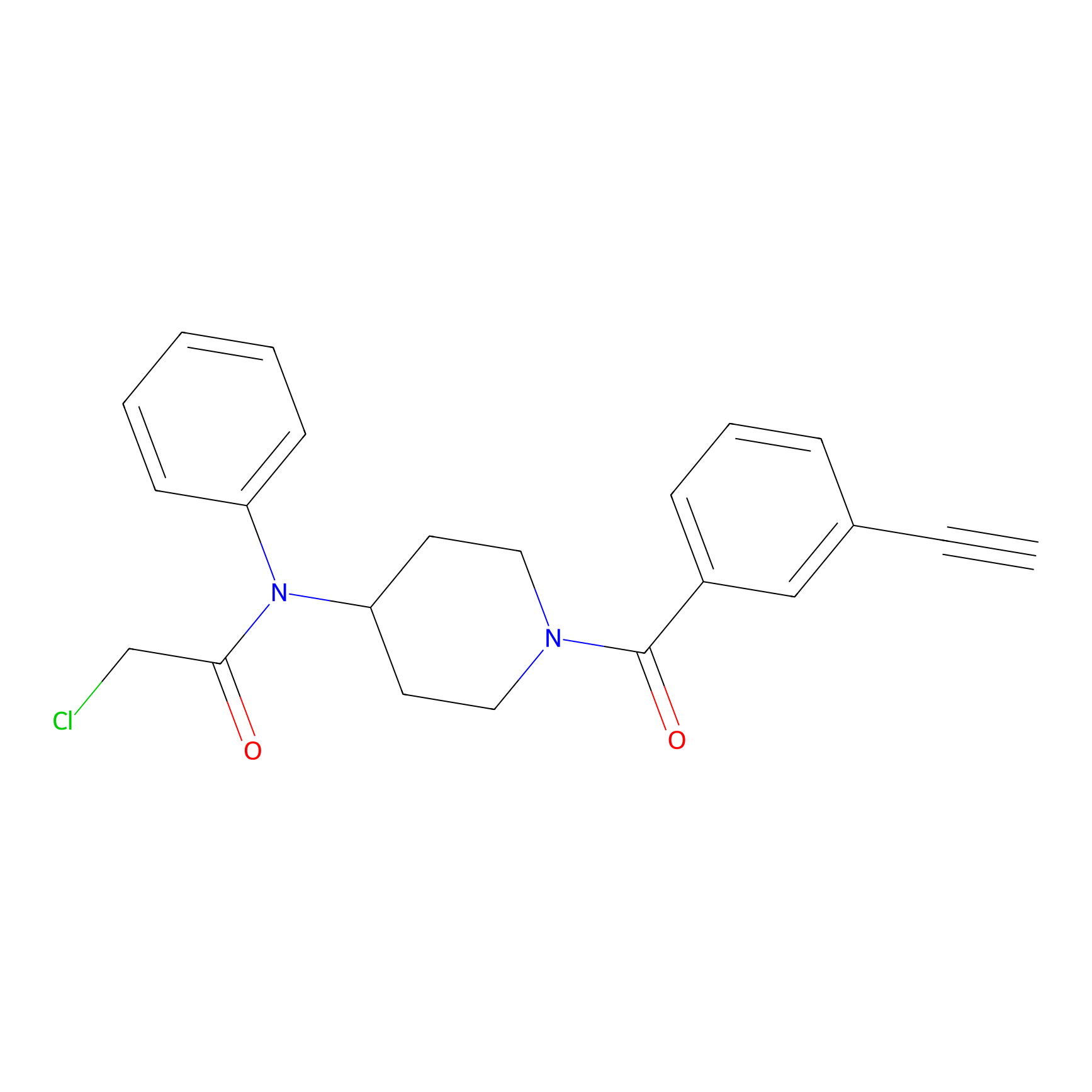 |
_(20.00) | LDD1485 | [6] | |
|
DA-P3 Probe Info |
 |
7.68 | LDD0180 | [7] | |
|
HHS-475 Probe Info |
 |
Y680(1.38) | LDD0264 | [8] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [9] | |
|
IPM Probe Info |
 |
C205(0.00); C296(0.00); C161(0.00) | LDD0025 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [10] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0151 | [11] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [12] | |
|
Phosphinate-6 Probe Info |
 |
C762(0.00); C205(0.00); C296(0.00) | LDD0018 | [13] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [14] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [14] | |
|
AOyne Probe Info |
 |
12.90 | LDD0443 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 11.65 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [14] |
| LDCM0028 | Dobutamine | HEK-293T | 7.68 | LDD0180 | [7] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C205(1.30); C296(1.08) | LDD1702 | [5] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 5.57 | LDD0183 | [7] |
| LDCM0625 | F8 | Ramos | C296(1.85); C205(2.19); C161(1.68) | LDD2187 | [16] |
| LDCM0572 | Fragment10 | Ramos | C296(1.80); C161(2.25) | LDD2189 | [16] |
| LDCM0573 | Fragment11 | Ramos | C296(0.36); C205(0.90); C161(0.20) | LDD2190 | [16] |
| LDCM0574 | Fragment12 | Ramos | C296(0.94); C205(0.89); C161(0.52) | LDD2191 | [16] |
| LDCM0575 | Fragment13 | Ramos | C296(0.77); C161(0.65) | LDD2192 | [16] |
| LDCM0576 | Fragment14 | Ramos | C296(0.36); C205(0.97); C161(0.54) | LDD2193 | [16] |
| LDCM0579 | Fragment20 | Ramos | C296(0.69); C205(0.81); C161(1.12) | LDD2194 | [16] |
| LDCM0580 | Fragment21 | Ramos | C296(0.81); C205(1.06); C161(1.02) | LDD2195 | [16] |
| LDCM0582 | Fragment23 | Ramos | C296(1.17) | LDD2196 | [16] |
| LDCM0578 | Fragment27 | Ramos | C296(1.02) | LDD2197 | [16] |
| LDCM0586 | Fragment28 | Ramos | C296(0.54); C205(0.56); C161(1.05) | LDD2198 | [16] |
| LDCM0588 | Fragment30 | Ramos | C296(0.95); C205(0.91); C161(1.46) | LDD2199 | [16] |
| LDCM0589 | Fragment31 | Ramos | C296(1.17); C205(1.22); C161(2.12) | LDD2200 | [16] |
| LDCM0590 | Fragment32 | Ramos | C296(3.23) | LDD2201 | [16] |
| LDCM0468 | Fragment33 | Ramos | C296(1.44); C205(2.02); C161(0.92) | LDD2202 | [16] |
| LDCM0596 | Fragment38 | Ramos | C296(1.23); C161(1.09) | LDD2203 | [16] |
| LDCM0566 | Fragment4 | Ramos | C296(1.14); C205(0.91); C161(1.05) | LDD2184 | [16] |
| LDCM0610 | Fragment52 | Ramos | C296(1.01); C161(0.69) | LDD2204 | [16] |
| LDCM0614 | Fragment56 | Ramos | C296(1.33); C205(1.44); C161(0.89) | LDD2205 | [16] |
| LDCM0569 | Fragment7 | Jurkat | _(20.00) | LDD1485 | [6] |
| LDCM0571 | Fragment9 | Ramos | C296(0.83); C205(0.73); C161(1.26) | LDD2188 | [16] |
| LDCM0116 | HHS-0101 | DM93 | Y680(1.38) | LDD0264 | [8] |
| LDCM0117 | HHS-0201 | DM93 | Y680(3.79) | LDD0265 | [8] |
| LDCM0118 | HHS-0301 | DM93 | Y680(4.06) | LDD0266 | [8] |
| LDCM0119 | HHS-0401 | DM93 | Y680(2.61) | LDD0267 | [8] |
| LDCM0120 | HHS-0701 | DM93 | Y680(1.67) | LDD0268 | [8] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [14] |
| LDCM0022 | KB02 | Ramos | C296(1.76); C205(1.88) | LDD2182 | [16] |
| LDCM0023 | KB03 | MDA-MB-231 | C205(1.96) | LDD1701 | [5] |
| LDCM0024 | KB05 | RD | C148(1.50) | LDD3401 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [14] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C296(1.48) | LDD2118 | [5] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C296(1.66) | LDD2124 | [5] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C296(0.80) | LDD2141 | [5] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C296(0.58) | LDD2150 | [5] |
| LDCM0029 | Quercetin | HEK-293T | 8.74 | LDD0181 | [7] |
| LDCM0170 | Structure8 | Ramos | 6.46 | LDD0433 | [17] |
References
