Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| 22RV1 | SNV: p.R56Q | . | |||
| HCT15 | SNV: p.S150R | DBIA Probe Info | |||
| HT115 | SNV: p.I147N | . | |||
| HUH7 | SNV: p.N35H | DBIA Probe Info | |||
| IM95 | SNV: p.L385P | DBIA Probe Info | |||
| MM1S | SNV: p.R452C | . | |||
| NCIH1793 | SNV: p.Y344Ter | DBIA Probe Info | |||
| NCIH1993 | SNV: p.G7S | . | |||
| SUPT1 | Deletion: p.G7VfsTer36 | DBIA Probe Info | |||
| TE1 | SNV: p.D113G | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.90 | LDD0402 | [1] | |
|
BTD Probe Info |
 |
C138(1.32) | LDD1699 | [2] | |
|
IPM Probe Info |
 |
C138(0.00); C205(0.00); C79(0.00); C71(0.00) | LDD0241 | [3] | |
|
Probe 1 Probe Info |
 |
Y501(7.84) | LDD3495 | [4] | |
|
HHS-475 Probe Info |
 |
Y63(0.65) | LDD0264 | [5] | |
|
DBIA Probe Info |
 |
C71(1.14); C72(1.14) | LDD0078 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
C79(0.00); C205(0.00); C429(0.00); C441(0.00) | LDD0036 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
C79(0.00); C429(0.00); C138(0.00); C205(0.00) | LDD0037 | [7] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [8] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [8] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [9] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [8] | |
|
W1 Probe Info |
 |
C138(0.00); C205(0.00) | LDD0236 | [3] | |
|
AOyne Probe Info |
 |
11.50 | LDD0443 | [10] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [11] | |
|
HHS-482 Probe Info |
 |
Y63(0.46) | LDD2239 | [12] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C087 Probe Info |
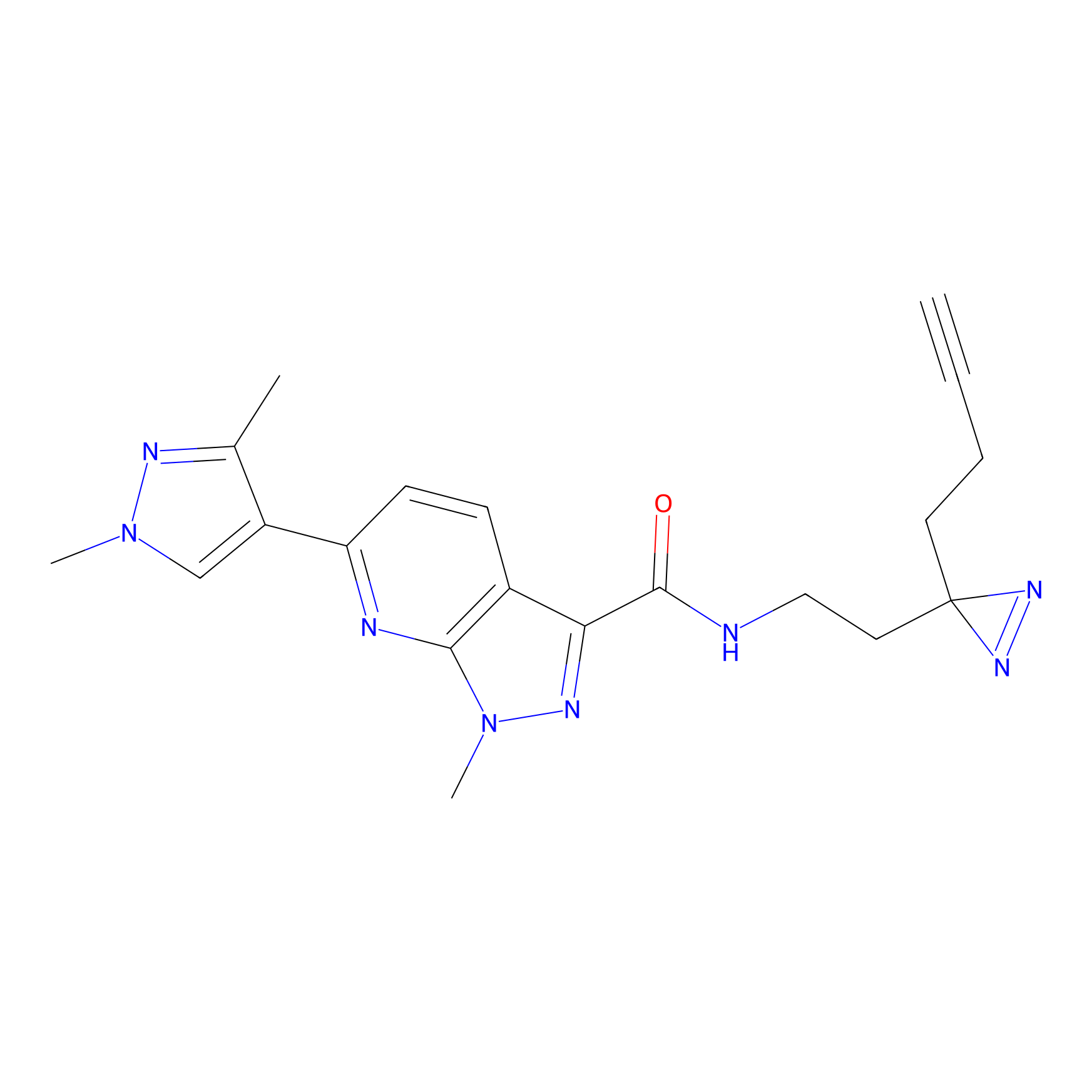 |
8.22 | LDD1779 | [13] | |
|
C129 Probe Info |
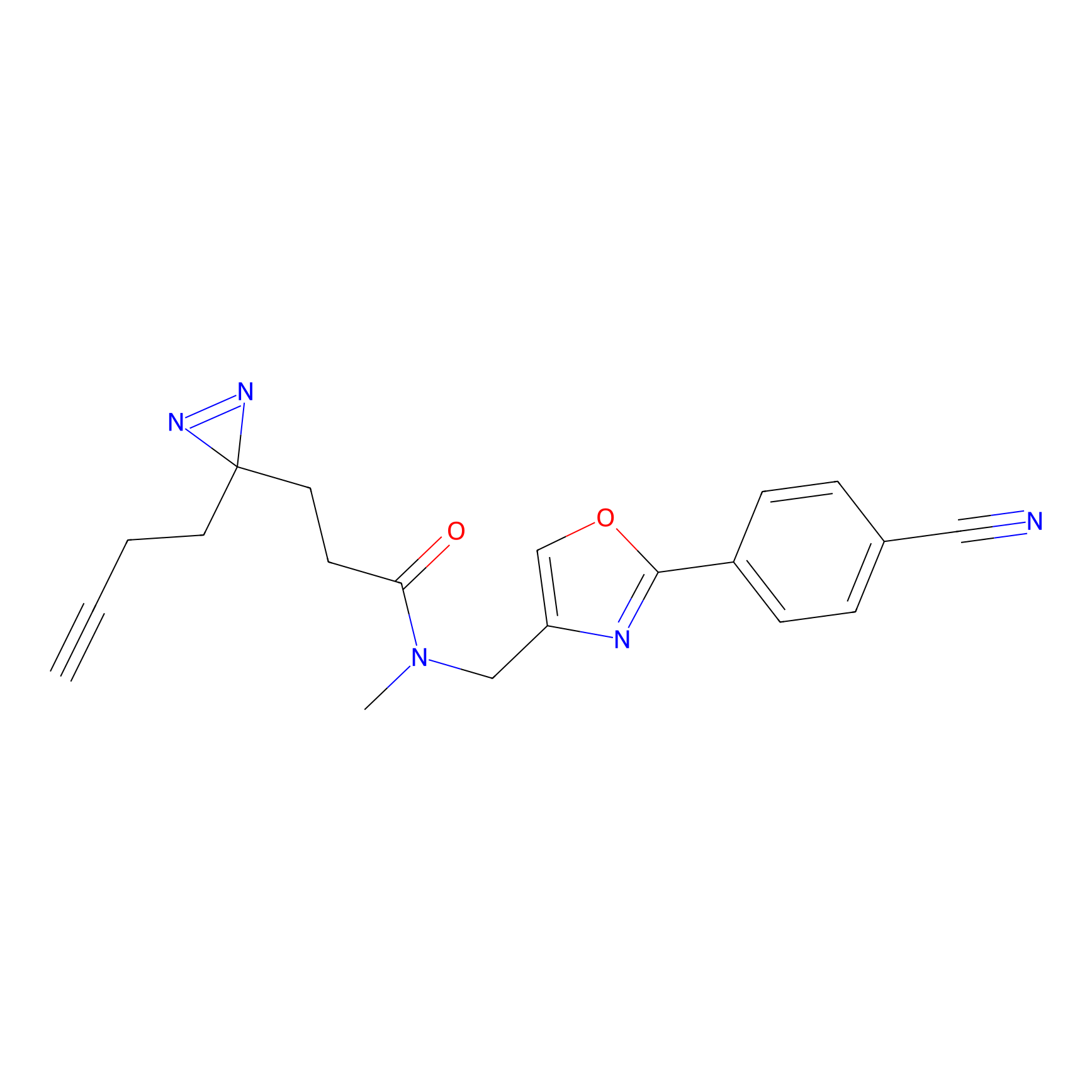 |
5.17 | LDD1811 | [13] | |
|
C165 Probe Info |
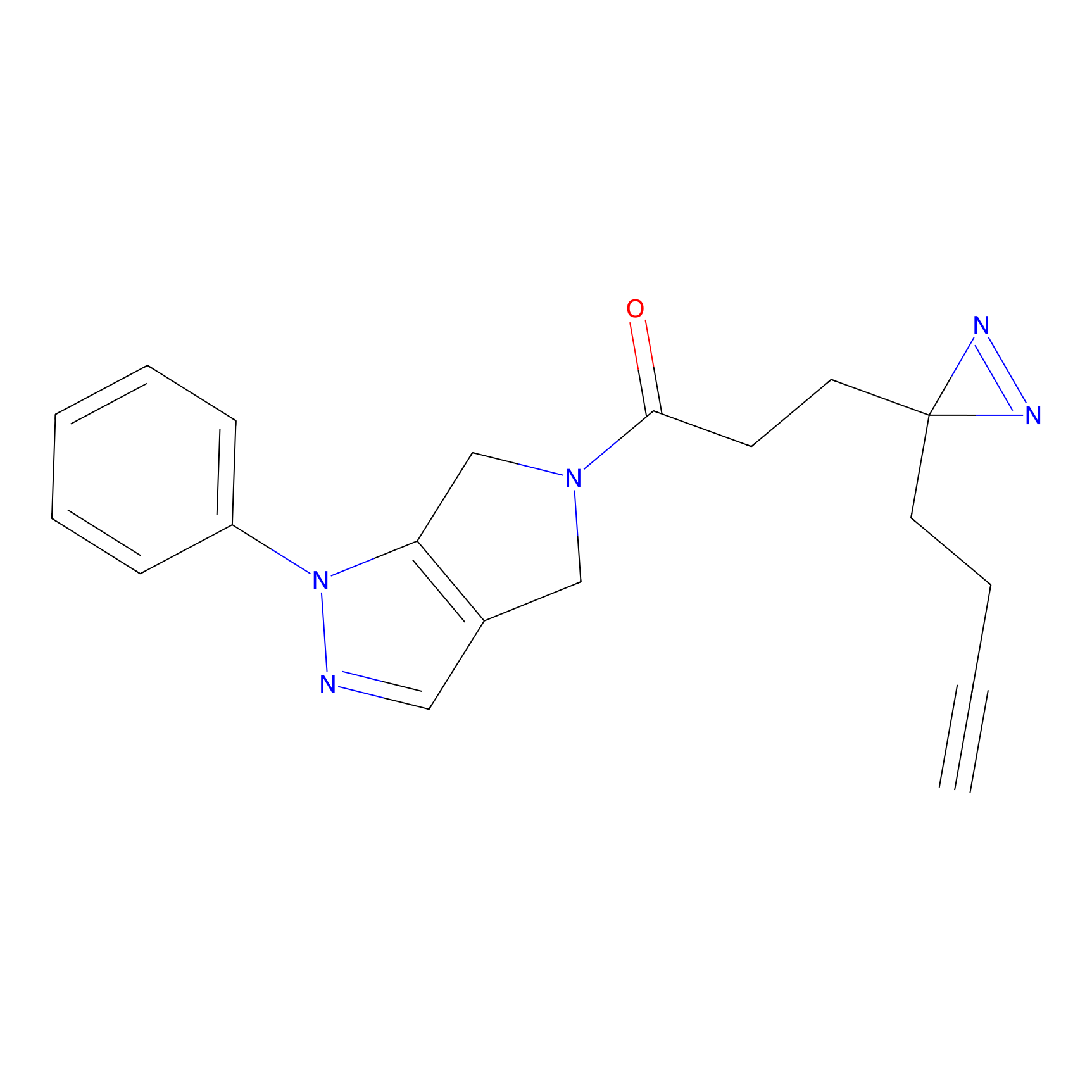 |
22.47 | LDD1845 | [13] | |
|
C166 Probe Info |
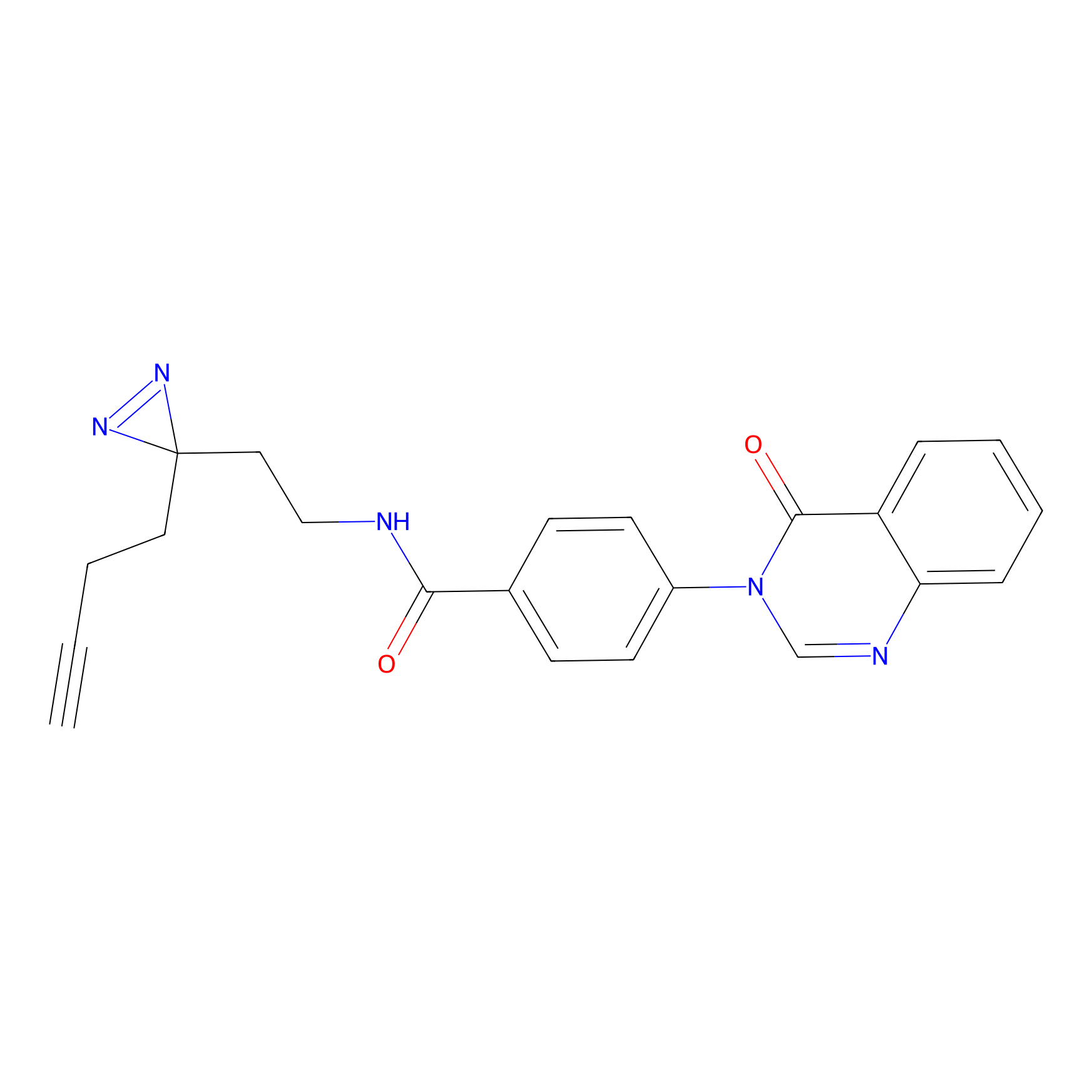 |
9.58 | LDD1846 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C205(0.93) | LDD2117 | [2] |
| LDCM0156 | Aniline | NCI-H1299 | 14.37 | LDD0403 | [1] |
| LDCM0020 | ARS-1620 | HCC44 | C71(1.14); C72(1.14) | LDD0078 | [6] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C205(1.18) | LDD2091 | [2] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C205(1.02) | LDD1702 | [2] |
| LDCM0116 | HHS-0101 | DM93 | Y63(0.65) | LDD0264 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y63(0.79) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y63(6.35) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y63(6.33) | LDD0268 | [5] |
| LDCM0022 | KB02 | 42-MG-BA | C310(1.41) | LDD2244 | [14] |
| LDCM0023 | KB03 | MDA-MB-231 | C205(1.32) | LDD1701 | [2] |
| LDCM0024 | KB05 | G361 | C79(1.67); C72(0.91) | LDD3311 | [14] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C205(0.41) | LDD2121 | [2] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C205(0.69) | LDD2098 | [2] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C205(1.15) | LDD2105 | [2] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C138(1.42) | LDD2107 | [2] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C205(0.91) | LDD2120 | [2] |
| LDCM0529 | Nucleophilic fragment 27b | MDA-MB-231 | C205(1.06) | LDD2122 | [2] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C205(1.31) | LDD2123 | [2] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C205(0.76) | LDD2127 | [2] |
| LDCM0535 | Nucleophilic fragment 30b | MDA-MB-231 | C205(1.11) | LDD2128 | [2] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C205(0.81) | LDD1700 | [2] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C205(0.71) | LDD2146 | [2] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C205(0.31) | LDD2150 | [2] |
The Interaction Atlas With This Target
References
