Details of the Target
General Information of Target
| Target ID | LDTP12285 | |||||
|---|---|---|---|---|---|---|
| Target Name | Charged multivesicular body protein 1a (CHMP1A) | |||||
| Gene Name | CHMP1A | |||||
| Gene ID | 5119 | |||||
| Synonyms |
CHMP1; KIAA0047; PCOLN3; PRSM1; Charged multivesicular body protein 1a; Chromatin-modifying protein 1a; CHMP1a; Vacuolar protein sorting-associated protein 46-1; Vps46-1; hVps46-1 |
|||||
| 3D Structure | ||||||
| Sequence |
MNRESFAAGERLVSPAYVRQGCEARRSHEHLIRLLLEKGKCPENGWDESTLELFLHELAI
MDSNNFLGNCGVGEREGRVASALVARRHYRFIHGIGRSGDISAVQPKAAGSSLLNKITNS LVLDIIKLAGVHTVANCFVVPMATGMSLTLCFLTLRHKRPKAKYIIWPRIDQKSCFKSMI TAGFEPVVIENVLEGDELRTDLKAVEAKVQELGPDCILCIHSTTSCFAPRVPDRLEELAV ICANYDIPHIVNNAYGVQSSKCMHLIQQGARVGRIDAFVQSLDKNFMVPVGGAIIAGFND SFIQEISKMYPGRASASPSLDVLITLLSLGSNGYKKLLKERKEMFSYLSNQIKKLSEAYN ERLLHTPHNPISLAMTLKTLDEHRDKAVTQLGSMLFTRQVSGARVVPLGSMQTVSGYTFR GFMSHTNNYPCAYLNAASAIGMKMQDVDLFIKRLDRCLKAVRKERSKESDDNYDKTEDVD IEEMALKLDNVLLDTYQDASS |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
SNF7 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Probable peripherally associated component of the endosomal sorting required for transport complex III (ESCRT-III) which is involved in multivesicular bodies (MVBs) formation and sorting of endosomal cargo proteins into MVBs. MVBs contain intraluminal vesicles (ILVs) that are generated by invagination and scission from the limiting membrane of the endosome and mostly are delivered to lysosomes enabling degradation of membrane proteins, such as stimulated growth factor receptors, lysosomal enzymes and lipids. The MVB pathway appears to require the sequential function of ESCRT-O, -I,-II and -III complexes. ESCRT-III proteins mostly dissociate from the invaginating membrane before the ILV is released. The ESCRT machinery also functions in topologically equivalent membrane fission events, such as the terminal stages of cytokinesis and the budding of enveloped viruses (HIV-1 and other lentiviruses). ESCRT-III proteins are believed to mediate the necessary vesicle extrusion and/or membrane fission activities, possibly in conjunction with the AAA ATPase VPS4. Involved in cytokinesis. Involved in recruiting VPS4A and/or VPS4B to the midbody of dividing cells. May also be involved in chromosome condensation. Targets the Polycomb group (PcG) protein BMI1/PCGF4 to regions of condensed chromatin. May play a role in stable cell cycle progression and in PcG gene silencing.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| KASUMI2 | SNV: p.K24Q | . | |||
| MOLT4 | SNV: p.D25N | IA-alkyne Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.93 | LDD0402 | [1] | |
|
STPyne Probe Info |
 |
K107(3.27); K13(6.20); K17(5.88); K83(4.72) | LDD0277 | [2] | |
|
BTD Probe Info |
 |
C44(6.50) | LDD1699 | [3] | |
|
Probe 1 Probe Info |
 |
Y48(57.70) | LDD3495 | [4] | |
|
DBIA Probe Info |
 |
C44(0.87) | LDD3316 | [5] | |
|
HHS-475 Probe Info |
 |
Y48(0.87) | LDD0264 | [6] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [7] | |
|
ATP probe Probe Info |
 |
K13(0.00); K40(0.00) | LDD0199 | [8] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [9] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [10] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [10] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [11] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [12] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [14] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [7] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [15] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [16] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C361 Probe Info |
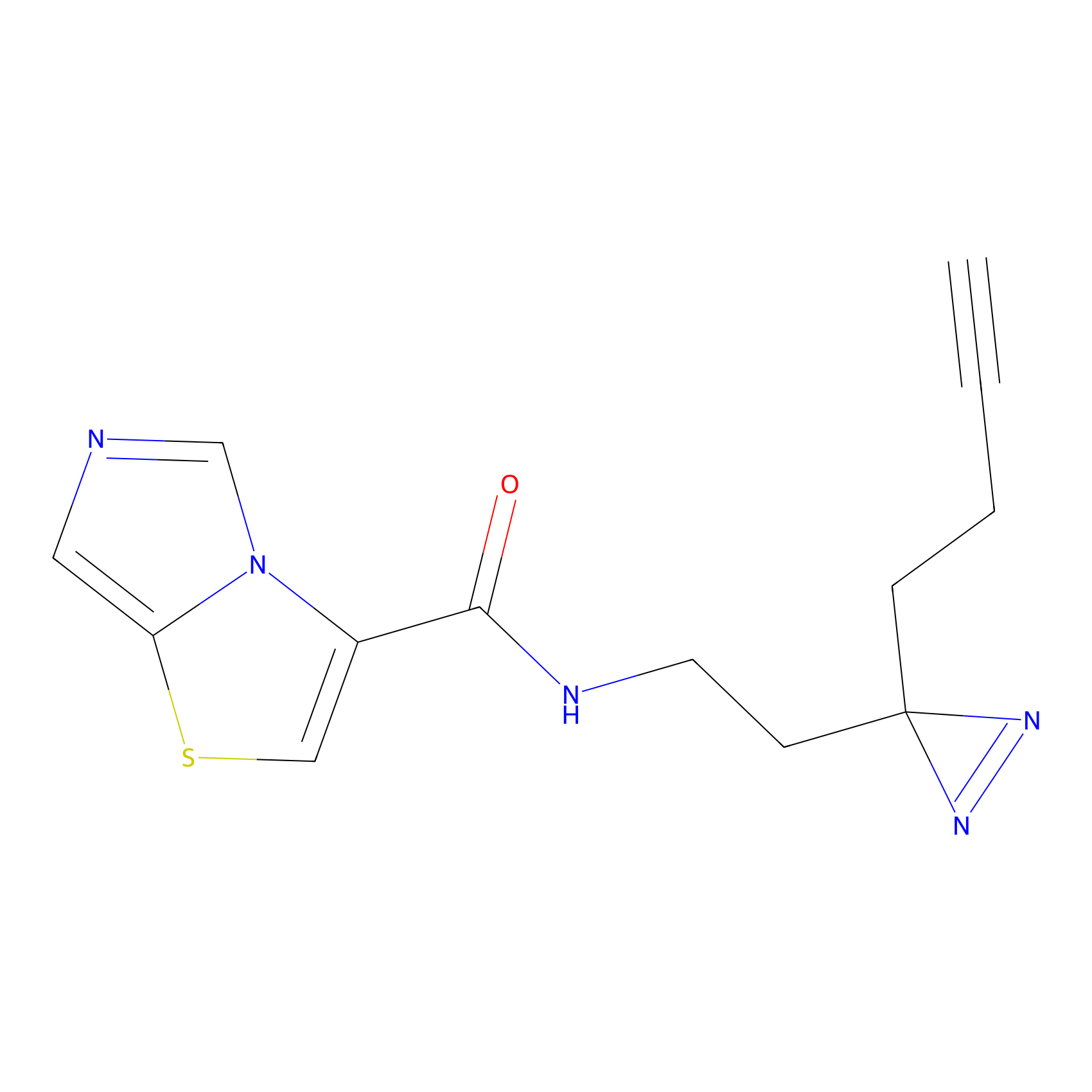 |
17.51 | LDD2022 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C44(1.07) | LDD2142 | [3] |
| LDCM0519 | 1-(6-methoxy-3,4-dihydroquinolin-1(2H)-yl)-2-nitroethan-1-one | MDA-MB-231 | C44(0.93) | LDD2112 | [3] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C44(1.00) | LDD2117 | [3] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C44(1.25) | LDD2152 | [3] |
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C44(0.86) | LDD2103 | [3] |
| LDCM0545 | Acetamide | MDA-MB-231 | C44(0.29) | LDD2138 | [3] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C44(0.63) | LDD2113 | [3] |
| LDCM0156 | Aniline | NCI-H1299 | 12.40 | LDD0403 | [1] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [7] |
| LDCM0198 | Dimethyl Fumarate(DMF) | T cell | C44(11.88) | LDD0513 | [18] |
| LDCM0116 | HHS-0101 | DM93 | Y48(0.87) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y48(0.83) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y48(0.91) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y48(0.86) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y48(0.81) | LDD0268 | [6] |
| LDCM0022 | KB02 | T cell | C44(5.12) | LDD1703 | [18] |
| LDCM0023 | KB03 | MDA-MB-231 | C44(1.71) | LDD1701 | [3] |
| LDCM0024 | KB05 | MEL167 | C44(0.87) | LDD3316 | [5] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C44(0.99) | LDD2102 | [3] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C44(0.57) | LDD2121 | [3] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C44(0.80) | LDD2089 | [3] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C44(0.77) | LDD2092 | [3] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C44(0.86) | LDD2093 | [3] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C44(0.71) | LDD2097 | [3] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C44(0.70) | LDD2099 | [3] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C44(1.00) | LDD2100 | [3] |
| LDCM0508 | Nucleophilic fragment 17a | MDA-MB-231 | C44(0.97) | LDD2101 | [3] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C44(0.86) | LDD2104 | [3] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C44(1.18) | LDD2105 | [3] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C44(0.81) | LDD2106 | [3] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C44(0.76) | LDD2107 | [3] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C44(0.66) | LDD2108 | [3] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C44(0.62) | LDD2109 | [3] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C44(0.82) | LDD2111 | [3] |
| LDCM0521 | Nucleophilic fragment 23b | MDA-MB-231 | C44(0.89) | LDD2114 | [3] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C44(0.41) | LDD2115 | [3] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C44(0.98) | LDD2120 | [3] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C44(0.61) | LDD2123 | [3] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C44(0.74) | LDD2125 | [3] |
| LDCM0533 | Nucleophilic fragment 29b | MDA-MB-231 | C44(1.92) | LDD2126 | [3] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C44(0.92) | LDD2127 | [3] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C44(0.53) | LDD2129 | [3] |
| LDCM0540 | Nucleophilic fragment 35 | MDA-MB-231 | C44(0.40) | LDD2133 | [3] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C44(0.39) | LDD2134 | [3] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C44(0.84) | LDD2136 | [3] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C44(0.88) | LDD2137 | [3] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C44(1.35) | LDD1700 | [3] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C44(0.70) | LDD2141 | [3] |
| LDCM0549 | Nucleophilic fragment 43 | MDA-MB-231 | C44(1.13) | LDD2143 | [3] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C44(4.36) | LDD2144 | [3] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C44(0.76) | LDD2146 | [3] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C44(0.43) | LDD2148 | [3] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C44(2.01) | LDD2153 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
| Alpha-synuclein (SNCA) | Synuclein family | P37840 | |||
| Syntenin-1 (SDCBP) | . | O00560 | |||
Other
References
