Details of the Target
General Information of Target
| Target ID | LDTP09948 | |||||
|---|---|---|---|---|---|---|
| Target Name | Peroxisomal membrane protein PEX13 (PEX13) | |||||
| Gene Name | PEX13 | |||||
| Gene ID | 5194 | |||||
| Synonyms |
Peroxisomal membrane protein PEX13; Peroxin-13 |
|||||
| 3D Structure | ||||||
| Sequence |
MAEGSRGGPTCSGVGGRQDPVSGSGGCNFPEYELPELNTRAFHVGAFGELWRGRLRGAGD
LSLREPPASALPGSQAADSDREDAAVARDLDCSLEAAAELRAVCGLDKLKCLEDGEDPEV IPENTDLVTLGVRKRFLEHREETITIDRACRQETFVYEMESHAIGKKPENSADMIEEGEL ILSVNILYPVIFHKHKEHKPYQTMLVLGSQKLTQLRDSIRCVSDLQIGGEFSNTPDQAPE HISKDLYKSAFFYFEGTFYNDKRYPECRDLSRTIIEWSESHDRGYGKFQTARMEDFTFND LCIKLGFPYLYCHQGDCEHVIVITDIRLVHHDDCLDRTLYPLLIKKHWLWTRKCFVCKMY TARWVTNNDSFAPEDPCFFCDVCFRMLHYDSEGNKLGEFLAYPYVDPGTFN |
|||||
| Target Bioclass |
Transporter and channel
|
|||||
| Family |
Peroxin-13 family
|
|||||
| Subcellular location |
Peroxisome membrane
|
|||||
| Function |
Component of the PEX13-PEX14 docking complex, a translocon channel that specifically mediates the import of peroxisomal cargo proteins bound to PEX5 receptor. The PEX13-PEX14 docking complex forms a large import pore which can be opened to a diameter of about 9 nm. Mechanistically, PEX5 receptor along with cargo proteins associates with the PEX14 subunit of the PEX13-PEX14 docking complex in the cytosol, leading to the insertion of the receptor into the organelle membrane with the concomitant translocation of the cargo into the peroxisome matrix. Involved in the import of PTS1- and PTS2-type containing proteins.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
JZ128-DTB Probe Info |
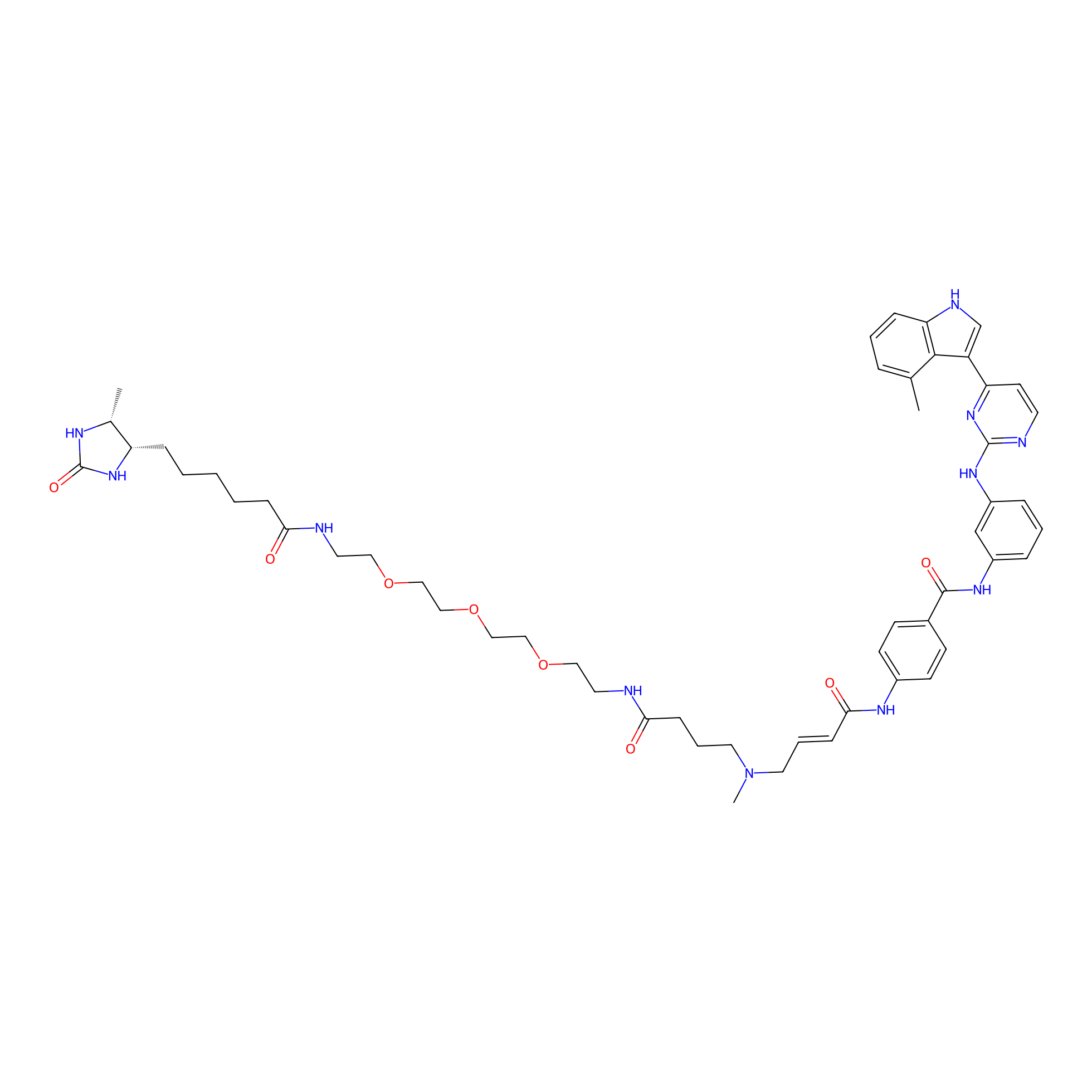 |
N.A. | LDD0462 | [2] | |
|
AHL-Pu-1 Probe Info |
 |
C220(2.20) | LDD0170 | [3] | |
|
IA-alkyne Probe Info |
 |
C220(20.00) | LDD1709 | [4] | |
|
DBIA Probe Info |
 |
C220(1.01) | LDD1512 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [6] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C289 Probe Info |
 |
35.02 | LDD1959 | [7] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | DM93 | C220(2.20) | LDD0170 | [3] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C220(4.07) | LDD0171 | [3] |
| LDCM0259 | AC14 | HEK-293T | C220(1.01) | LDD1512 | [5] |
| LDCM0282 | AC22 | HEK-293T | C220(1.47) | LDD1521 | [5] |
| LDCM0291 | AC30 | HEK-293T | C220(1.24) | LDD1530 | [5] |
| LDCM0299 | AC38 | HEK-293T | C220(0.93) | LDD1538 | [5] |
| LDCM0308 | AC46 | HEK-293T | C220(1.04) | LDD1547 | [5] |
| LDCM0317 | AC54 | HEK-293T | C220(1.11) | LDD1556 | [5] |
| LDCM0323 | AC6 | HEK-293T | C220(1.01) | LDD1562 | [5] |
| LDCM0326 | AC62 | HEK-293T | C220(1.01) | LDD1565 | [5] |
| LDCM0368 | CL10 | HEK-293T | C220(1.42) | LDD1572 | [5] |
| LDCM0410 | CL22 | HEK-293T | C220(1.36) | LDD1614 | [5] |
| LDCM0423 | CL34 | HEK-293T | C220(1.15) | LDD1627 | [5] |
| LDCM0436 | CL46 | HEK-293T | C220(0.93) | LDD1640 | [5] |
| LDCM0449 | CL58 | HEK-293T | C220(1.10) | LDD1652 | [5] |
| LDCM0463 | CL70 | HEK-293T | C220(1.54) | LDD1666 | [5] |
| LDCM0476 | CL82 | HEK-293T | C220(1.17) | LDD1679 | [5] |
| LDCM0489 | CL94 | HEK-293T | C220(0.88) | LDD1692 | [5] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [2] |
| LDCM0024 | KB05 | T cell | C220(20.00) | LDD1709 | [4] |
The Interaction Atlas With This Target
References
