Details of the Target
General Information of Target
| Target ID | LDTP09895 | |||||
|---|---|---|---|---|---|---|
| Target Name | Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1 (INPP5D) | |||||
| Gene Name | INPP5D | |||||
| Gene ID | 3635 | |||||
| Synonyms |
SHIP; SHIP1; Phosphatidylinositol 3,4,5-trisphosphate 5-phosphatase 1; EC 3.1.3.86; Inositol polyphosphate-5-phosphatase D; EC 3.1.3.56; Inositol polyphosphate-5-phosphatase of 145 kDa; SIP-145; Phosphatidylinositol 4,5-bisphosphate 5-phosphatase; EC 3.1.3.36; SH2 domain-containing inositol 5'-phosphatase 1; SH2 domain-containing inositol phosphatase 1; SHIP-1; p150Ship; hp51CN
|
|||||
| 3D Structure | ||||||
| Sequence |
MREPEELMPDSGAVFTFGKSKFAENNPGKFWFKNDVPVHLSCGDEHSAVVTGNNKLYMFG
SNNWGQLGLGSKSAISKPTCVKALKPEKVKLAACGRNHTLVSTEGGNVYATGGNNEGQLG LGDTEERNTFHVISFFTSEHKIKQLSAGSNTSAALTEDGRLFMWGDNSEGQIGLKNVSNV CVPQQVTIGKPVSWISCGYYHSAFVTTDGELYVFGEPENGKLGLPNQLLGNHRTPQLVSE IPEKVIQVACGGEHTVVLTENAVYTFGLGQFGQLGLGTFLFETSEPKVIENIRDQTISYI SCGENHTALITDIGLMYTFGDGRHGKLGLGLENFTNHFIPTLCSNFLRFIVKLVACGGCH MVVFAAPHRGVAKEIEFDEINDTCLSVATFLPYSSLTSGNVLQRTLSARMRRRERERSPD SFSMRRTLPPIEGTLGLSACFLPNSVFPRCSERNLQESVLSEQDLMQPEEPDYLLDEMTK EAEIDNSSTVESLGETTDILNMTHIMSLNSNEKSLKLSPVQKQKKQQTIGELTQDTALTE NDDSDEYEEMSEMKEGKACKQHVSQGIFMTQPATTIEAFSDEEVGNDTGQVGPQADTDGE GLQKEVYRHENNNGVDQLDAKEIEKESDGGHSQKESEAEEIDSEKETKLAEIAGMKDLRE REKSTKKMSPFFGNLPDRGMNTESEENKDFVKKRESCKQDVIFDSERESVEKPDSYMEGA SESQQGIADGFQQPEAIEFSSGEKEDDEVETDQNIRYGRKLIEQGNEKETKPIISKSMAK YDFKCDRLSEIPEEKEGAEDSKGNGIEEQEVEANEENVKVHGGRKEKTEILSDDLTDKAE DHEFSKTEELKLEDVDEEINAENVESKKKTVGDDESVPTGYHSKTEGAERTNDDSSAETI EKKEKANLEERAICEYNENPKGYMLDDADSSSLEILENSETTPSKDMKKTKKIFLFKRVP SINQKIVKNNNEPLPEIKSIGDQIILKSDNKDADQNHMSQNHQNIPPTNTERRSKSCTIL |
|||||
| Target Type |
Clinical trial
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Inositol 1,4,5-trisphosphate 5-phosphatase family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Phosphatidylinositol (PtdIns) phosphatase that specifically hydrolyzes the 5-phosphate of phosphatidylinositol-3,4,5-trisphosphate (PtdIns(3,4,5)P3) to produce PtdIns(3,4)P2, thereby negatively regulating the PI3K (phosphoinositide 3-kinase) pathways. Able also to hydrolyzes the 5-phosphate of phosphatidylinositol-4,5-bisphosphate (PtdIns(4,5)P3) and inositol 1,3,4,5-tetrakisphosphate. Acts as a negative regulator of B-cell antigen receptor signaling. Mediates signaling from the FC-gamma-RIIB receptor (FCGR2B), playing a central role in terminating signal transduction from activating immune/hematopoietic cell receptor systems. Acts as a negative regulator of myeloid cell proliferation/survival and chemotaxis, mast cell degranulation, immune cells homeostasis, integrin alpha-IIb/beta-3 signaling in platelets and JNK signaling in B-cells. Regulates proliferation of osteoclast precursors, macrophage programming, phagocytosis and activation and is required for endotoxin tolerance. Involved in the control of cell-cell junctions, CD32a signaling in neutrophils and modulation of EGF-induced phospholipase C activity. Key regulator of neutrophil migration, by governing the formation of the leading edge and polarization required for chemotaxis. Modulates FCGR3/CD16-mediated cytotoxicity in NK cells. Mediates the activin/TGF-beta-induced apoptosis through its Smad-dependent expression.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| 786O | SNV: p.G922R | . | |||
| A101D | SNV: p.H753Y | . | |||
| AGS | SNV: p.K337T | DBIA Probe Info | |||
| DU145 | SNV: p.D239E; p.G1171V | . | |||
| FTC133 | SNV: p.T1136P | . | |||
| HCC15 | SNV: p.E751D | . | |||
| HEC1 | SNV: p.R1140W | . | |||
| JHH7 | SNV: p.R1162H | . | |||
| KPNYN | SNV: p.T1136P | . | |||
| NB1 | SNV: p.D436E | . | |||
| NCIH23 | SNV: p.P1058S | DBIA Probe Info | |||
| OPM2 | Deletion: p.P1180_P1182del | DBIA Probe Info | |||
| SNU1 | Deletion: p.E775SfsTer4 | . | |||
| TOV21G | SNV: p.E139Ter; p.G919W | . | |||
| U937 | SNV: p.A1080T | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
IPM Probe Info |
 |
C1088(0.00); C506(0.00) | LDD0241 | [1] | |
|
DBIA Probe Info |
 |
C506(1.36) | LDD3314 | [2] | |
|
Alkyne-RA190 Probe Info |
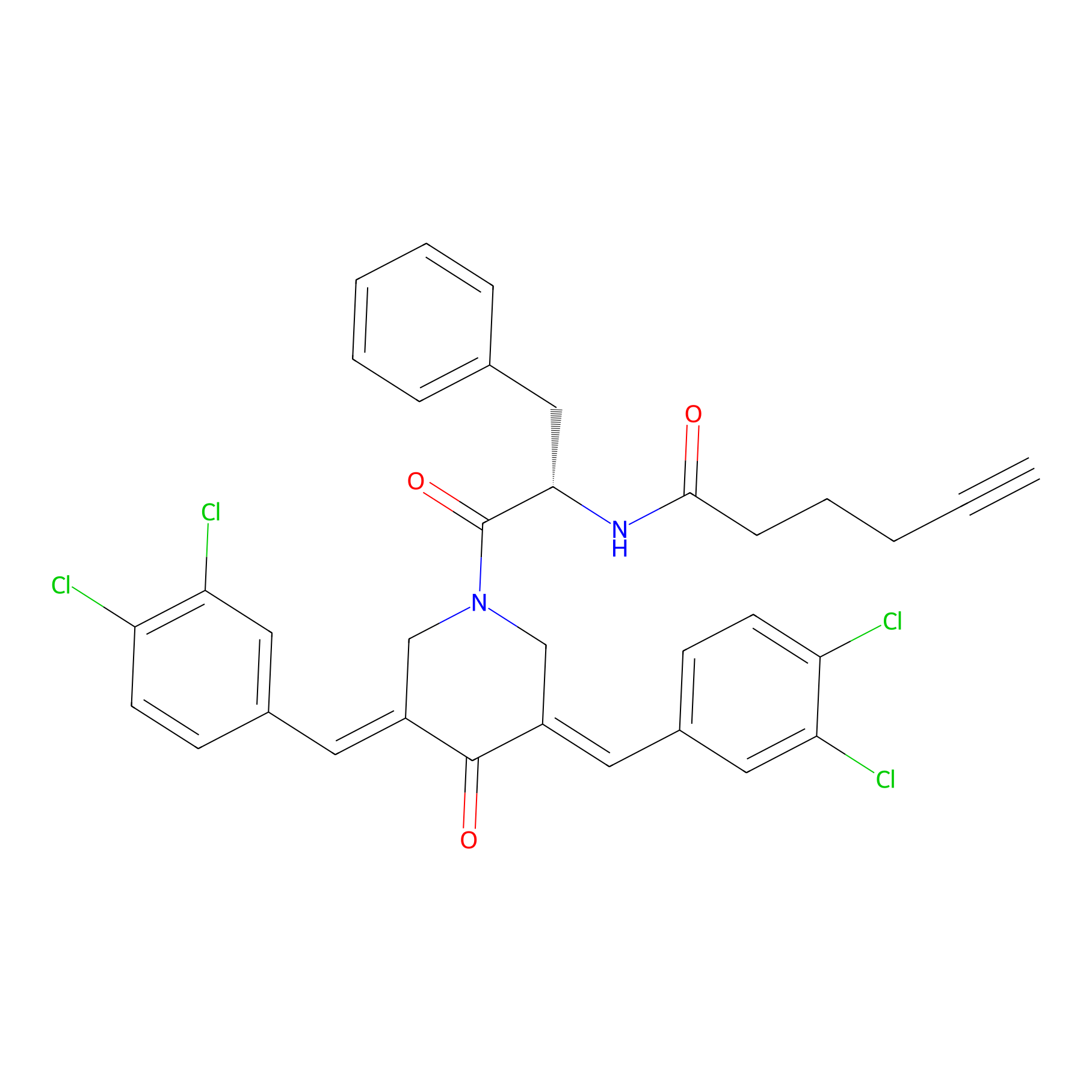 |
3.35 | LDD0300 | [3] | |
|
W1 Probe Info |
 |
C1088(0.86) | LDD0239 | [1] | |
|
IA-alkyne Probe Info |
 |
C506(0.00); C385(0.00); C672(0.00) | LDD0167 | [4] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [5] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0198 | Dimethyl Fumarate(DMF) | T cell | C1088(5.11) | LDD0513 | [6] |
| LDCM0625 | F8 | Ramos | C506(1.44); C1088(1.03); C138(1.47); C217(0.75) | LDD2187 | [7] |
| LDCM0572 | Fragment10 | Ramos | C506(0.70); C1088(0.75); C138(1.21); C217(1.46) | LDD2189 | [7] |
| LDCM0573 | Fragment11 | Ramos | C506(5.92); C1088(3.25); C138(0.31); C217(16.72) | LDD2190 | [7] |
| LDCM0574 | Fragment12 | Ramos | C506(0.65); C1088(0.31); C138(0.93); C217(2.02) | LDD2191 | [7] |
| LDCM0575 | Fragment13 | Ramos | C506(1.09); C1088(0.79); C138(0.99); C217(0.38) | LDD2192 | [7] |
| LDCM0576 | Fragment14 | Ramos | C506(1.31); C1088(1.28); C138(0.80); C217(0.86) | LDD2193 | [7] |
| LDCM0579 | Fragment20 | Ramos | C506(0.52); C1088(0.34); C138(0.90); C217(1.48) | LDD2194 | [7] |
| LDCM0580 | Fragment21 | Ramos | C506(0.86); C1088(1.05); C138(1.02); C217(0.68) | LDD2195 | [7] |
| LDCM0582 | Fragment23 | Ramos | C506(0.66); C1088(0.51); C138(0.96); C217(2.84) | LDD2196 | [7] |
| LDCM0578 | Fragment27 | Ramos | C506(0.88); C1088(0.91); C138(1.09); C217(0.89) | LDD2197 | [7] |
| LDCM0586 | Fragment28 | Ramos | C506(0.93); C1088(0.59); C138(1.18); C217(0.50) | LDD2198 | [7] |
| LDCM0588 | Fragment30 | Ramos | C506(0.99); C1088(0.61); C138(1.55); C217(1.34) | LDD2199 | [7] |
| LDCM0589 | Fragment31 | Ramos | C506(0.79); C1088(0.44); C138(1.11); C217(2.34) | LDD2200 | [7] |
| LDCM0590 | Fragment32 | Ramos | C506(0.67); C1088(0.81); C138(1.36); C217(2.99) | LDD2201 | [7] |
| LDCM0468 | Fragment33 | Ramos | C506(1.07); C1088(0.63); C138(1.23); C217(1.79) | LDD2202 | [7] |
| LDCM0596 | Fragment38 | Ramos | C506(1.00); C1088(0.69); C138(0.95); C217(0.48) | LDD2203 | [7] |
| LDCM0566 | Fragment4 | Ramos | C506(1.22); C1088(0.96); C138(0.96); C217(0.84) | LDD2184 | [7] |
| LDCM0610 | Fragment52 | Ramos | C506(1.39); C1088(0.90); C138(1.28); C217(0.60) | LDD2204 | [7] |
| LDCM0614 | Fragment56 | Ramos | C506(1.19); C1088(0.50); C138(1.37); C217(0.48) | LDD2205 | [7] |
| LDCM0569 | Fragment7 | Ramos | C506(0.87); C1088(0.83); C138(1.19); C217(0.74) | LDD2186 | [7] |
| LDCM0571 | Fragment9 | Ramos | C506(0.79); C1088(0.44); C138(0.99); C217(2.37) | LDD2188 | [7] |
| LDCM0022 | KB02 | Ramos | C506(1.30); C1088(1.66); C138(1.35); C217(0.87) | LDD2182 | [7] |
| LDCM0023 | KB03 | Ramos | C506(1.15); C1088(1.55); C138(1.03); C217(2.29) | LDD2183 | [7] |
| LDCM0024 | KB05 | IGR37 | C506(1.36) | LDD3314 | [2] |
| LDCM0131 | RA190 | MM1.R | 3.35 | LDD0300 | [3] |
| LDCM0170 | Structure8 | Ramos | 5.13 | LDD0433 | [8] |
| LDCM0112 | W16 | Hep-G2 | C1088(0.86) | LDD0239 | [1] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Immunoglobulin
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Low affinity immunoglobulin gamma Fc region receptor II-b (FCGR2B) | . | P31994 | |||
| Natural killer cell receptor 2B4 (CD244) | . | Q9BZW8 | |||
Other
References
