Details of the Target
General Information of Target
| Target ID | LDTP05507 | |||||
|---|---|---|---|---|---|---|
| Target Name | Cyclin-dependent kinase 18 (CDK18) | |||||
| Gene Name | CDK18 | |||||
| Gene ID | 5129 | |||||
| Synonyms |
PCTAIRE3; PCTK3; Cyclin-dependent kinase 18; EC 2.7.11.22; Cell division protein kinase 18; PCTAIRE-motif protein kinase 3; Serine/threonine-protein kinase PCTAIRE-3 |
|||||
| 3D Structure | ||||||
| Sequence |
MIMNKMKNFKRRFSLSVPRTETIEESLAEFTEQFNQLHNRRNENLQLGPLGRDPPQECST
FSPTDSGEEPGQLSPGVQFQRRQNQRRFSMEDVSKRLSLPMDIRLPQEFLQKLQMESPDL PKPLSRMSRRASLSDIGFGKLETYVKLDKLGEGTYATVFKGRSKLTENLVALKEIRLEHE EGAPCTAIREVSLLKNLKHANIVTLHDLIHTDRSLTLVFEYLDSDLKQYLDHCGNLMSMH NVKIFMFQLLRGLAYCHHRKILHRDLKPQNLLINERGELKLADFGLARAKSVPTKTYSNE VVTLWYRPPDVLLGSTEYSTPIDMWGVGCIHYEMATGRPLFPGSTVKEELHLIFRLLGTP TEETWPGVTAFSEFRTYSFPCYLPQPLINHAPRLDTDGIHLLSSLLLYESKSRMSAEAAL SHSYFRSLGERVHQLEDTASIFSLKEIQLQKDPGYRGLAFQQPGRGKNRRQSIF |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein kinase superfamily, CMGC Ser/Thr protein kinase family, CDC2/CDKX subfamily
|
|||||
| Function | May play a role in signal transduction cascades in terminally differentiated cells. | |||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K173(10.00) | LDD0277 | [1] | |
|
DBIA Probe Info |
 |
C411(1.69) | LDD3344 | [2] | |
|
BTD Probe Info |
 |
C185(0.94) | LDD2090 | [3] | |
|
Johansson_61 Probe Info |
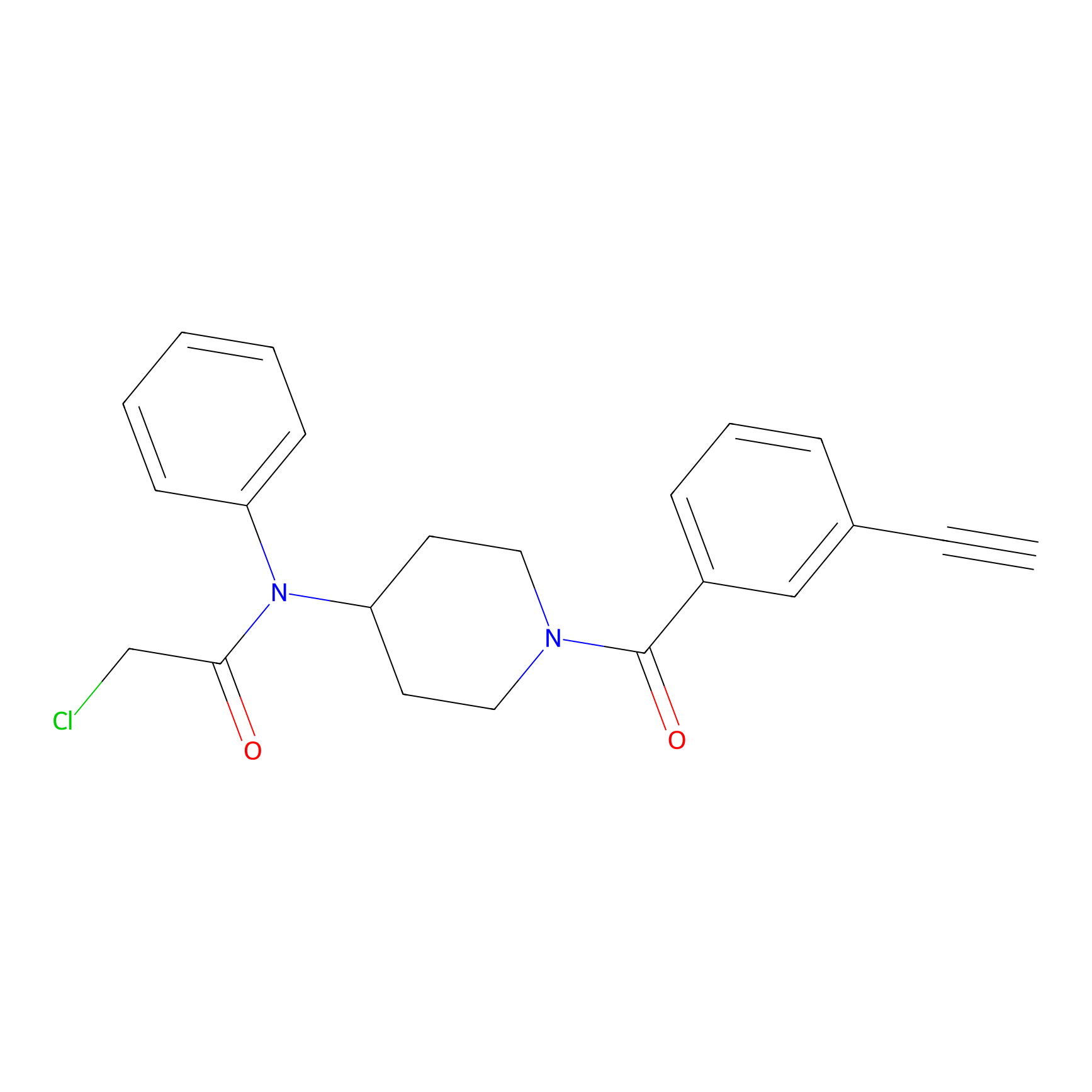 |
_(20.00) | LDD1485 | [4] | |
|
AHL-Pu-1 Probe Info |
 |
C183(3.04) | LDD0169 | [5] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C185(0.00); C381(0.00) | LDD0038 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0162 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
C185(0.00); C381(0.00) | LDD0037 | [6] | |
|
NAIA_5 Probe Info |
 |
C185(0.00); C206(0.00); C233(0.00) | LDD2224 | [8] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [9] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [9] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [9] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [9] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [10] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [9] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [11] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [12] | |
|
HHS-465 Probe Info |
 |
Y155(0.00); K160(0.00) | LDD2240 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DA-2 Probe Info |
 |
N.A. | LDD0070 | [14] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0510 | 3-(4-(Hydroxydiphenylmethyl)piperidin-1-yl)-3-oxopropanenitrile | MDA-MB-231 | C185(1.30) | LDD2103 | [3] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C183(3.04) | LDD0169 | [5] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C185(0.57) | LDD2091 | [3] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [12] |
| LDCM0632 | CL-Sc | Hep-G2 | C185(1.80); C381(1.75) | LDD2227 | [8] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C185(6.51) | LDD1702 | [3] |
| LDCM0569 | Fragment7 | Jurkat | _(20.00) | LDD1485 | [4] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [12] |
| LDCM0022 | KB02 | 769-P | C411(1.37) | LDD2246 | [2] |
| LDCM0023 | KB03 | 769-P | C411(2.01) | LDD2663 | [2] |
| LDCM0024 | KB05 | NCI-H146 | C411(1.69) | LDD3344 | [2] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C185(0.89) | LDD2121 | [3] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C185(0.94) | LDD2090 | [3] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C185(0.86) | LDD2092 | [3] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C185(0.89) | LDD2111 | [3] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C185(0.87) | LDD2140 | [3] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C185(0.49) | LDD2150 | [3] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C185(0.07) | LDD2206 | [16] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C185(0.70) | LDD2207 | [16] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| 14-3-3 protein zeta/delta (YWHAZ) | 14-3-3 family | P63104 | |||
Transcription factor
Other
References
