Details of the Target
General Information of Target
| Target ID | LDTP05436 | |||||
|---|---|---|---|---|---|---|
| Target Name | Aldo-keto reductase family 1 member C1 (AKR1C1) | |||||
| Gene Name | AKR1C1 | |||||
| Gene ID | 1645 | |||||
| Synonyms |
DDH; DDH1; Aldo-keto reductase family 1 member C1; EC 1.1.1.-; EC 1.1.1.112; EC 1.1.1.209; EC 1.1.1.210; EC 1.1.1.357; EC 1.1.1.51; EC 1.1.1.53; EC 1.1.1.62; EC 1.3.1.20; 20-alpha-hydroxysteroid dehydrogenase; 20-alpha-HSD; EC 1.1.1.149; Chlordecone reductase homolog HAKRC; Dihydrodiol dehydrogenase 1; DD1; High-affinity hepatic bile acid-binding protein; HBAB
|
|||||
| 3D Structure | ||||||
| Sequence |
MDSKYQCVKLNDGHFMPVLGFGTYAPAEVPKSKALEATKLAIEAGFRHIDSAHLYNNEEQ
VGLAIRSKIADGSVKREDIFYTSKLWCNSHRPELVRPALERSLKNLQLDYVDLYLIHFPV SVKPGEEVIPKDENGKILFDTVDLCATWEAVEKCKDAGLAKSIGVSNFNRRQLEMILNKP GLKYKPVCNQVECHPYFNQRKLLDFCKSKDIVLVAYSALGSHREEPWVDPNSPVLLEDPV LCALAKKHKRTPALIALRYQLQRGVVVLAKSYNEQRIRQNVQVFEFQLTSEEMKAIDGLN RNVRYLTLDIFAGPPNYPFSDEY |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Aldo/keto reductase family
|
|||||
| Subcellular location |
Cytoplasm, cytosol
|
|||||
| Function |
Cytosolic aldo-keto reductase that catalyzes the NADH and NADPH-dependent reduction of ketosteroids to hydroxysteroids. Most probably acts as a reductase in vivo since the oxidase activity measured in vitro is inhibited by physiological concentrations of NADPH. Displays a broad positional specificity acting on positions 3, 17 and 20 of steroids and regulates the metabolism of hormones like estrogens and androgens. May also reduce conjugated steroids such as 5alpha-dihydrotestosterone sulfate. Displays affinity for bile acids.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [1] | |
|
YN-4 Probe Info |
 |
100.00 | LDD0445 | [1] | |
|
STPyne Probe Info |
 |
K31(0.52); K33(5.01); K39(8.41) | LDD0277 | [2] | |
|
IPM Probe Info |
 |
C87(0.00); C145(0.00) | LDD0241 | [3] | |
|
BTD Probe Info |
 |
C193(0.79) | LDD2123 | [4] | |
|
NAIA_5 Probe Info |
 |
C7(0.76) | LDD2227 | [5] | |
|
Acrolein Probe Info |
 |
C87(0.00); H90(0.00) | LDD0221 | [6] | |
|
DBIA Probe Info |
 |
C188(0.90); C193(0.90) | LDD0078 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [8] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [9] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0149 | [10] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [6] | |
|
W1 Probe Info |
 |
N.A. | LDD0236 | [3] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [11] | |
|
HHS-475 Probe Info |
 |
Y55(1.73) | LDD2238 | [12] | |
|
HHS-482 Probe Info |
 |
Y55(1.14) | LDD2239 | [12] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe11 Probe Info |
 |
7.01 | LDD0472 | [13] | |
|
STS-2 Probe Info |
 |
2.86 | LDD0138 | [14] | |
|
AEA-DA Probe Info |
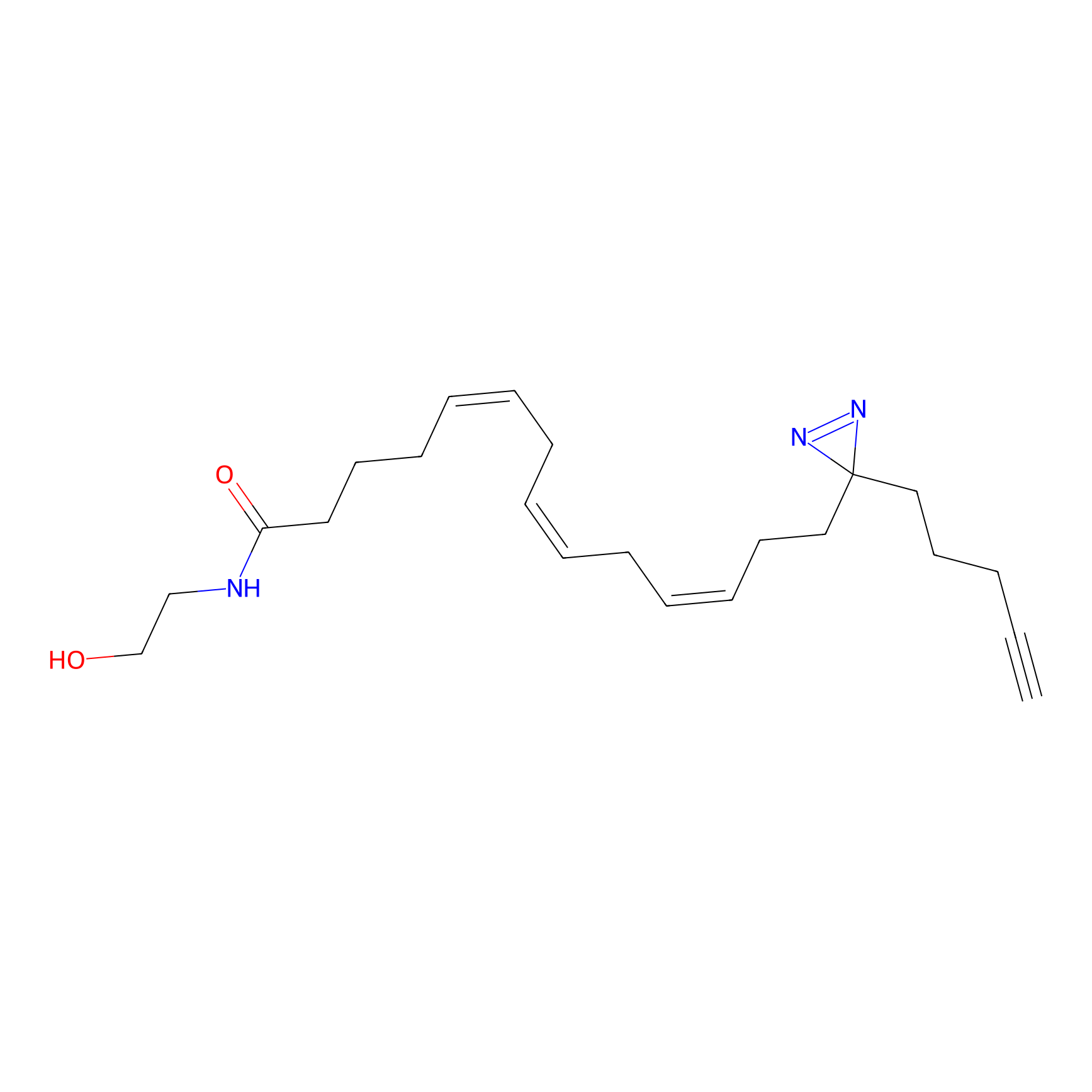 |
9.33 | LDD0146 | [15] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0071 | [16] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0068 | [17] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0020 | ARS-1620 | HCC44 | C188(0.90); C193(0.90) | LDD0078 | [7] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [6] |
| LDCM0632 | CL-Sc | Hep-G2 | C7(0.76) | LDD2227 | [5] |
| LDCM0082 | FK866 | A-549 | 9.33 | LDD0146 | [15] |
| LDCM0107 | IAA | HeLa | C87(0.00); H90(0.00) | LDD0221 | [6] |
| LDCM0022 | KB02 | 42-MG-BA | C145(1.23) | LDD2244 | [18] |
| LDCM0023 | KB03 | 42-MG-BA | C145(1.17); C87(2.13) | LDD2661 | [18] |
| LDCM0024 | KB05 | MEL167 | C87(1.38) | LDD3316 | [18] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C193(0.79) | LDD2123 | [4] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C188(0.76) | LDD2127 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C188(0.80) | LDD2136 | [4] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C193(0.87) | LDD2137 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Tyrosine-protein phosphatase non-receptor type 3 (PTPN3) | Protein-tyrosine phosphatase family | P26045 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Sprouty-related, EVH1 domain-containing protein 1 (SPRED1) | . | Q7Z699 | |||
The Drug(s) Related To This Target
Approved
Investigative
Discontinued
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Hexestrol | Small molecular drug | DB07931 | |||
References
