Details of the Target
General Information of Target
| Target ID | LDTP04543 | |||||
|---|---|---|---|---|---|---|
| Target Name | Aldo-keto reductase family 1 member C2 (AKR1C2) | |||||
| Gene Name | AKR1C2 | |||||
| Gene ID | 1646 | |||||
| Synonyms |
DDH2; Aldo-keto reductase family 1 member C2; EC 1.-.-.-; EC 1.1.1.112; EC 1.1.1.209; EC 1.1.1.53; EC 1.1.1.62; EC 1.3.1.20; 3-alpha-HSD3; Chlordecone reductase homolog HAKRD; Dihydrodiol dehydrogenase 2; DD-2; DD2; Dihydrodiol dehydrogenase/bile acid-binding protein; DD/BABP; Type III 3-alpha-hydroxysteroid dehydrogenase; EC 1.1.1.357
|
|||||
| 3D Structure | ||||||
| Sequence |
MDSKYQCVKLNDGHFMPVLGFGTYAPAEVPKSKALEAVKLAIEAGFHHIDSAHVYNNEEQ
VGLAIRSKIADGSVKREDIFYTSKLWSNSHRPELVRPALERSLKNLQLDYVDLYLIHFPV SVKPGEEVIPKDENGKILFDTVDLCATWEAMEKCKDAGLAKSIGVSNFNHRLLEMILNKP GLKYKPVCNQVECHPYFNQRKLLDFCKSKDIVLVAYSALGSHREEPWVDPNSPVLLEDPV LCALAKKHKRTPALIALRYQLQRGVVVLAKSYNEQRIRQNVQVFEFQLTSEEMKAIDGLN RNVRYLTLDIFAGPPNYPFSDEY |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Aldo/keto reductase family
|
|||||
| Subcellular location |
Cytoplasm, cytosol
|
|||||
| Function |
Cytosolic aldo-keto reductase that catalyzes the NADH and NADPH-dependent reduction of ketosteroids to hydroxysteroids. Most probably acts as a reductase in vivo since the oxidase activity measured in vitro is inhibited by physiological concentrations of NADPH. Displays a broad positional specificity acting on positions 3, 17 and 20 of steroids and regulates the metabolism of hormones like estrogens and androgens. Works in concert with the 5-alpha/5-beta-steroid reductases to convert steroid hormones into the 3-alpha/5-alpha and 3-alpha/5-beta-tetrahydrosteroids. Catalyzes the inactivation of the most potent androgen 5-alpha-dihydrotestosterone (5-alpha-DHT) to 5-alpha-androstane-3-alpha,17-beta-diol (3-alpha-diol). Also specifically able to produce 17beta-hydroxy-5alpha-androstan-3-one/5alphaDHT. May also reduce conjugated steroids such as 5alpha-dihydrotestosterone sulfate. Displays affinity for bile acids.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K161(7.69) | LDD0277 | [1] | |
|
DBIA Probe Info |
 |
C242(0.85) | LDD3312 | [2] | |
|
BTD Probe Info |
 |
C188(1.02) | LDD2093 | [3] | |
|
5E-2FA Probe Info |
 |
H14(0.00); H170(0.00) | LDD2235 | [4] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
C154(0.00); C188(0.00); C193(0.00); C7(0.00) | LDD0162 | [6] | |
|
IPM Probe Info |
 |
C188(0.00); C193(0.00) | LDD0025 | [7] | |
|
JW-RF-010 Probe Info |
 |
C7(0.00); C188(0.00); C193(0.00) | LDD0026 | [7] | |
|
TFBX Probe Info |
 |
C7(0.00); C188(0.00); C206(0.00); C193(0.00) | LDD0027 | [7] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [8] | |
|
Acrolein Probe Info |
 |
H14(0.00); H170(0.00) | LDD0217 | [9] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [9] | |
|
Methacrolein Probe Info |
 |
C188(0.00); C193(0.00) | LDD0218 | [9] | |
|
W1 Probe Info |
 |
C193(0.00); C188(0.00); D12(0.00); Y24(0.00) | LDD0236 | [10] | |
|
NAIA_5 Probe Info |
 |
C145(0.00); C242(0.00); C206(0.00); C7(0.00) | LDD2223 | [11] | |
|
HHS-465 Probe Info |
 |
N.A. | LDD2240 | [12] | |
|
HHS-475 Probe Info |
 |
Y110(0.70); Y114(1.61); Y184(1.43); Y196(1.27) | LDD2238 | [13] | |
|
HHS-482 Probe Info |
 |
Y110(1.46); Y114(1.53); Y184(1.02); Y196(0.85) | LDD2239 | [13] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STS-1 Probe Info |
 |
3.89 | LDD0136 | [14] | |
|
STS-2 Probe Info |
 |
6.44 | LDD0138 | [14] | |
|
AEA-DA Probe Info |
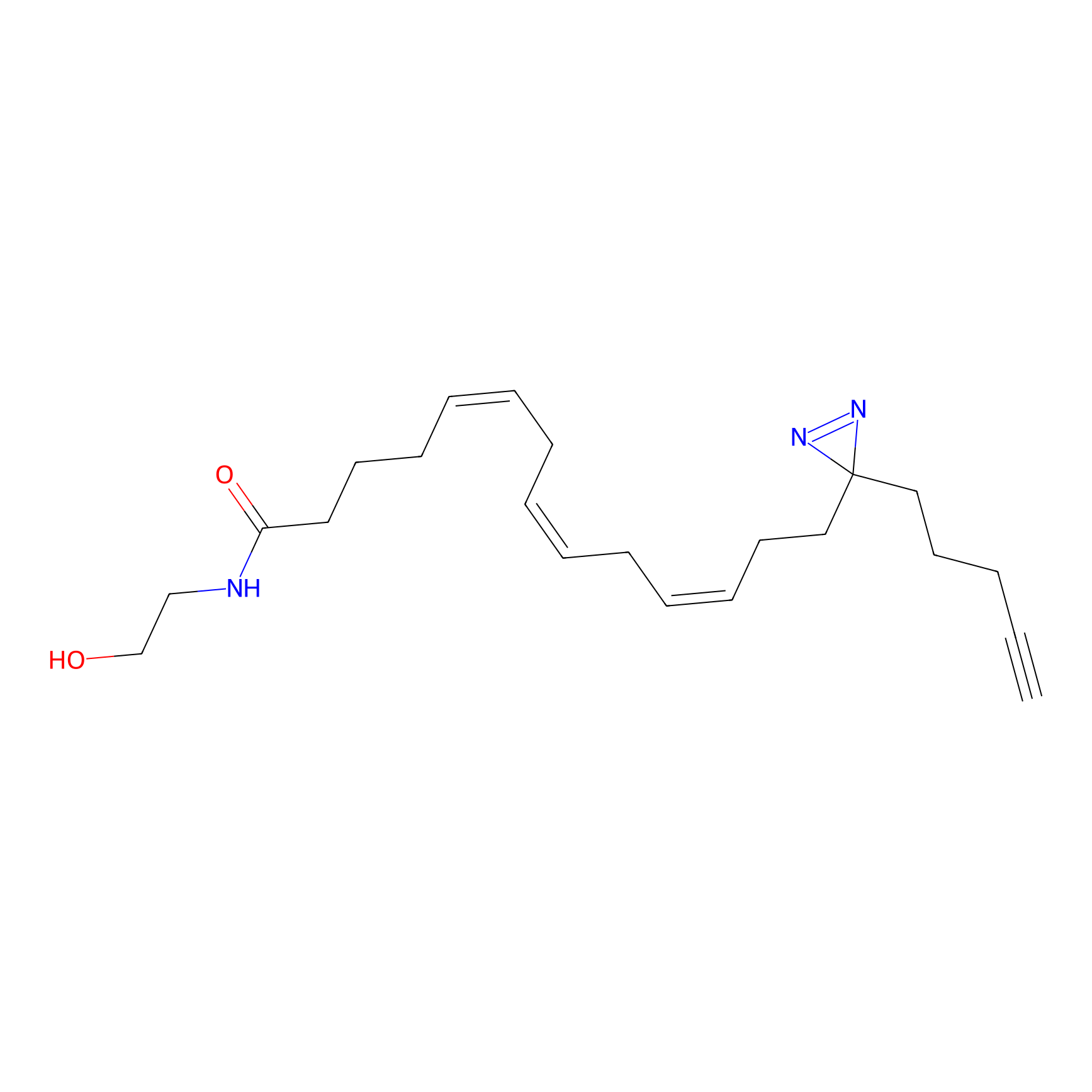 |
9.20 | LDD0146 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | H14(0.00); H170(0.00); H90(0.00) | LDD0222 | [9] |
| LDCM0082 | FK866 | A-549 | 9.20 | LDD0146 | [15] |
| LDCM0107 | IAA | HeLa | H14(0.00); H170(0.00); H90(0.00) | LDD0221 | [9] |
| LDCM0022 | KB02 | 22RV1 | C242(0.67) | LDD2243 | [2] |
| LDCM0023 | KB03 | 22RV1 | C242(0.68) | LDD2660 | [2] |
| LDCM0024 | KB05 | HMCB | C242(0.85) | LDD3312 | [2] |
| LDCM0109 | NEM | HeLa | H14(0.00); H90(0.00); H170(0.00) | LDD0223 | [9] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C188(1.02) | LDD2093 | [3] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C193(0.95) | LDD2119 | [3] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C188(0.90); C193(0.81) | LDD2123 | [3] |
| LDCM0110 | W12 | Hep-G2 | K201(1.26); K209(2.12); S208(2.12) | LDD0237 | [10] |
| LDCM0111 | W14 | Hep-G2 | K185(0.73); S208(1.75); K209(1.78); S32(1.78) | LDD0238 | [10] |
| LDCM0112 | W16 | Hep-G2 | K185(0.98); K179(1.24); N178(1.24); Y184(1.24) | LDD0239 | [10] |
| LDCM0113 | W17 | Hep-G2 | K185(0.60); Y184(0.60); K209(1.24); S208(1.24) | LDD0240 | [10] |
The Interaction Atlas With This Target
The Drug(s) Related To This Target
Approved
Investigative
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Epitestosterone | . | DB07768 | |||
| Nicotinamide Adenine Dinucleotide Phosphate | . | DB03461 | |||
References
