Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
HDSF-alk Probe Info |
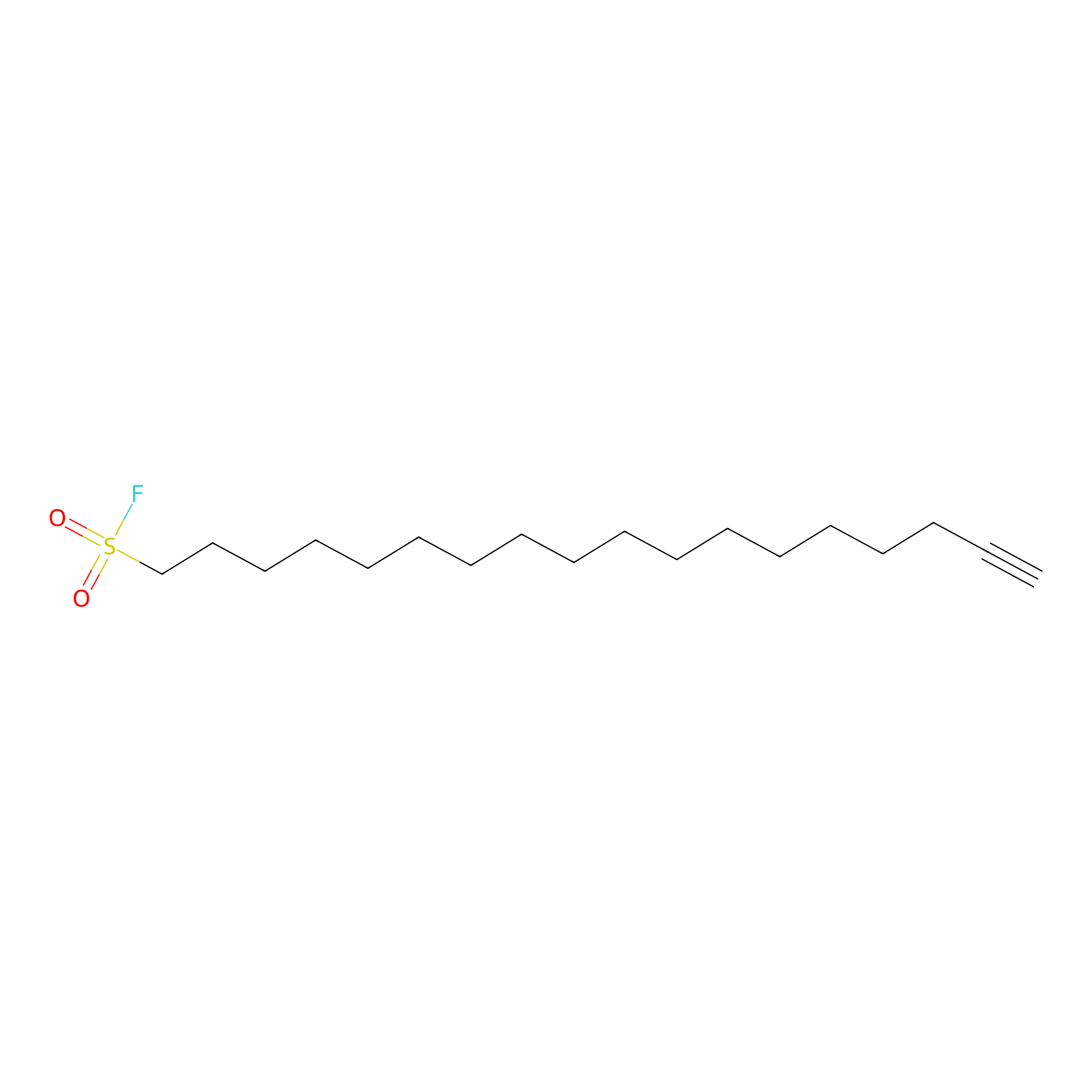 |
2.61 | LDD0197 | [1] | |
|
FBPP2 Probe Info |
 |
30.38 | LDD0318 | [2] | |
|
CY-1 Probe Info |
 |
100.00 | LDD0243 | [3] | |
|
CY4 Probe Info |
 |
100.00 | LDD0244 | [3] | |
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [4] | |
|
OPA-S-S-alkyne Probe Info |
 |
K194(2.39); K162(4.92); K110(5.07) | LDD3494 | [5] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [6] | |
|
AOyne Probe Info |
 |
5.00 | LDD0443 | [7] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [9] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [9] | |
|
LD-F Probe Info |
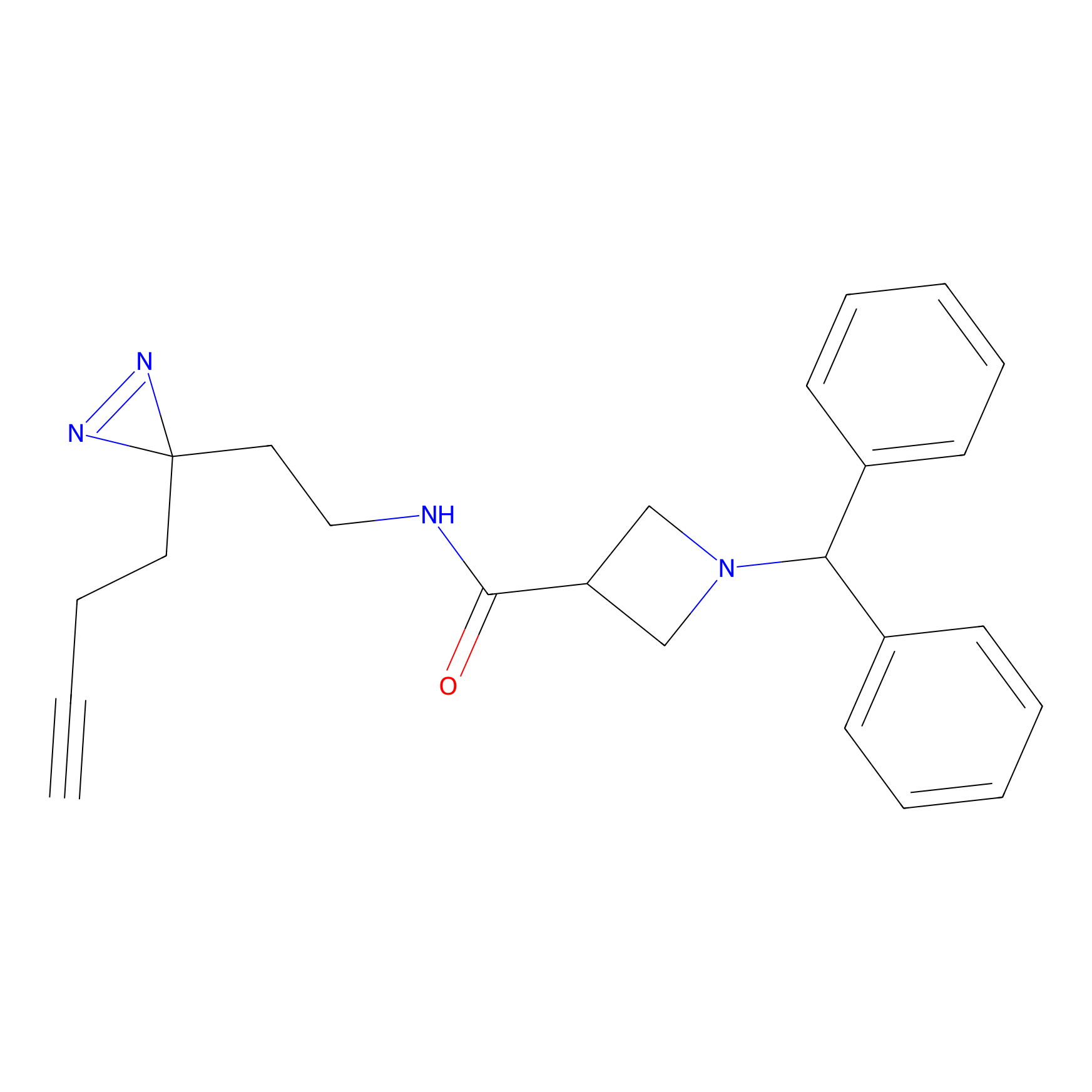 |
N.A. | LDD0015 | [10] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
References
