Details of the Target
General Information of Target
| Target ID | LDTP01914 | |||||
|---|---|---|---|---|---|---|
| Target Name | Putative HLA class I histocompatibility antigen, alpha chain H (HLA-H) | |||||
| Gene Name | HLA-H | |||||
| Synonyms |
HLAH; Putative HLA class I histocompatibility antigen, alpha chain H; HLA-12.4; HLA-AR; MHC class I antigen H |
|||||
| 3D Structure | ||||||
| Sequence |
MVLMAPRTLLLLLSGALALTQTWARSHSMRYFYTTMSRPGAGEPRFISVGYVDDTQFVRF
DSDDASPREEPRAPWMEREGPKYWDRNTQICKAQAQTERENLRIALRYYNQSEGGSHTMQ VMYGCDVGPDGPFLRGYEQHAYDGKDYIALNEDLRSWTAADMAAQITKRKWEAARRAEQR RVYLEGEFVEWLRRYLENGKETLQRADPPKTHMTHHPISDHEATLRCWALGFYPAEITLT WQRDGEDQTQDTELVETRPAGDGTFQKWAAVVVPSGEEQRYTCHVQHEGLPEPLTLRWEP SSQPTVPIVGIVAGLVLLVAVVTGAVVAAVMWRKKSSDRKGGSYSQAASSNSAQGSDVSL TA |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
MHC class I family
|
|||||
| Subcellular location |
Cell membrane
|
|||||
| Function | Involved in the presentation of foreign antigens to the immune system. | |||||
| Uniprot ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
P3 Probe Info |
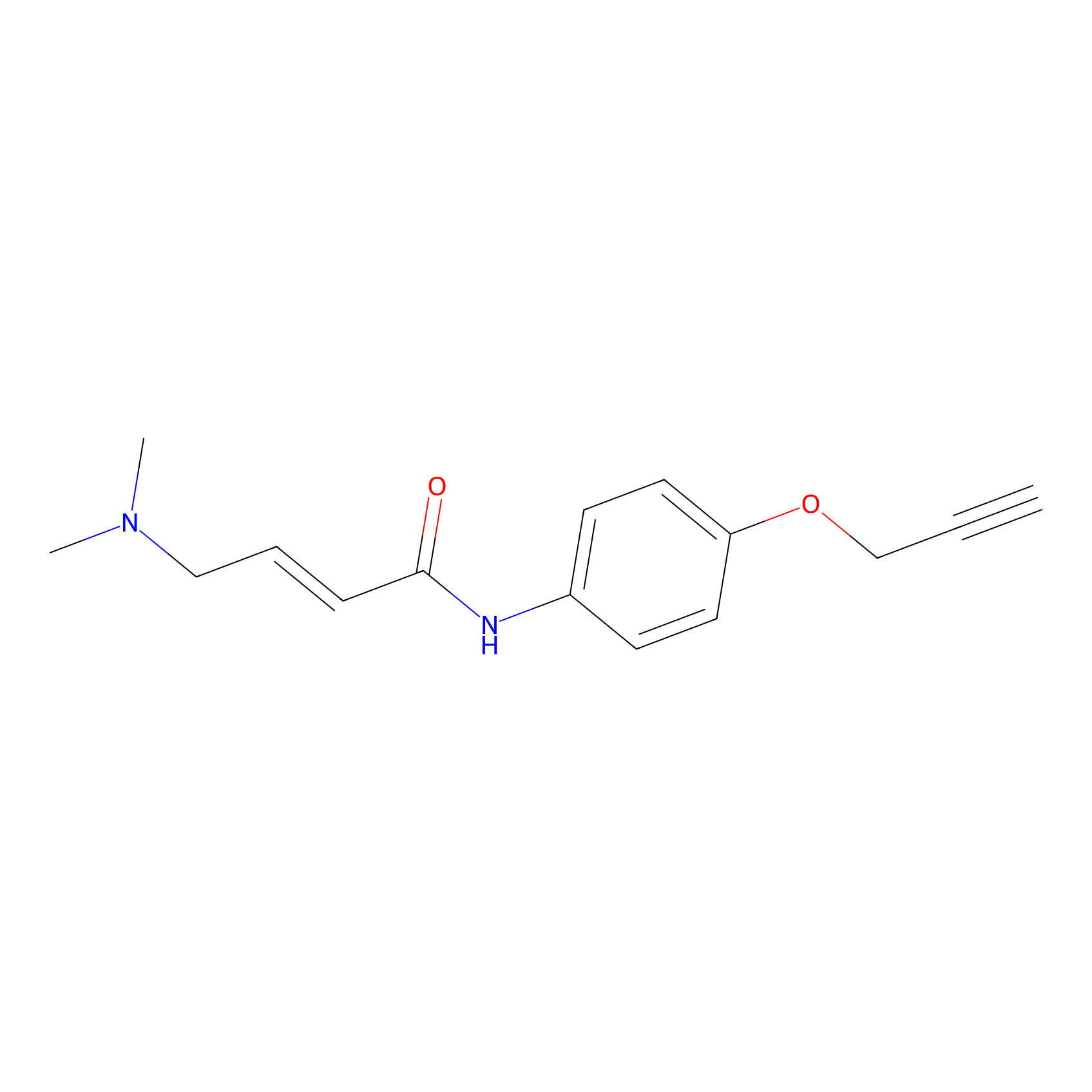 |
10.00 | LDD0450 | [2] | |
|
P8 Probe Info |
 |
10.00 | LDD0451 | [2] | |
|
IPM Probe Info |
 |
N.A. | LDD0241 | [3] | |
|
OPA-S-S-alkyne Probe Info |
 |
K200(1.46); K210(2.51) | LDD3494 | [4] | |
|
DBIA Probe Info |
 |
C283(2.03) | LDD3310 | [5] | |
|
YY4-yne Probe Info |
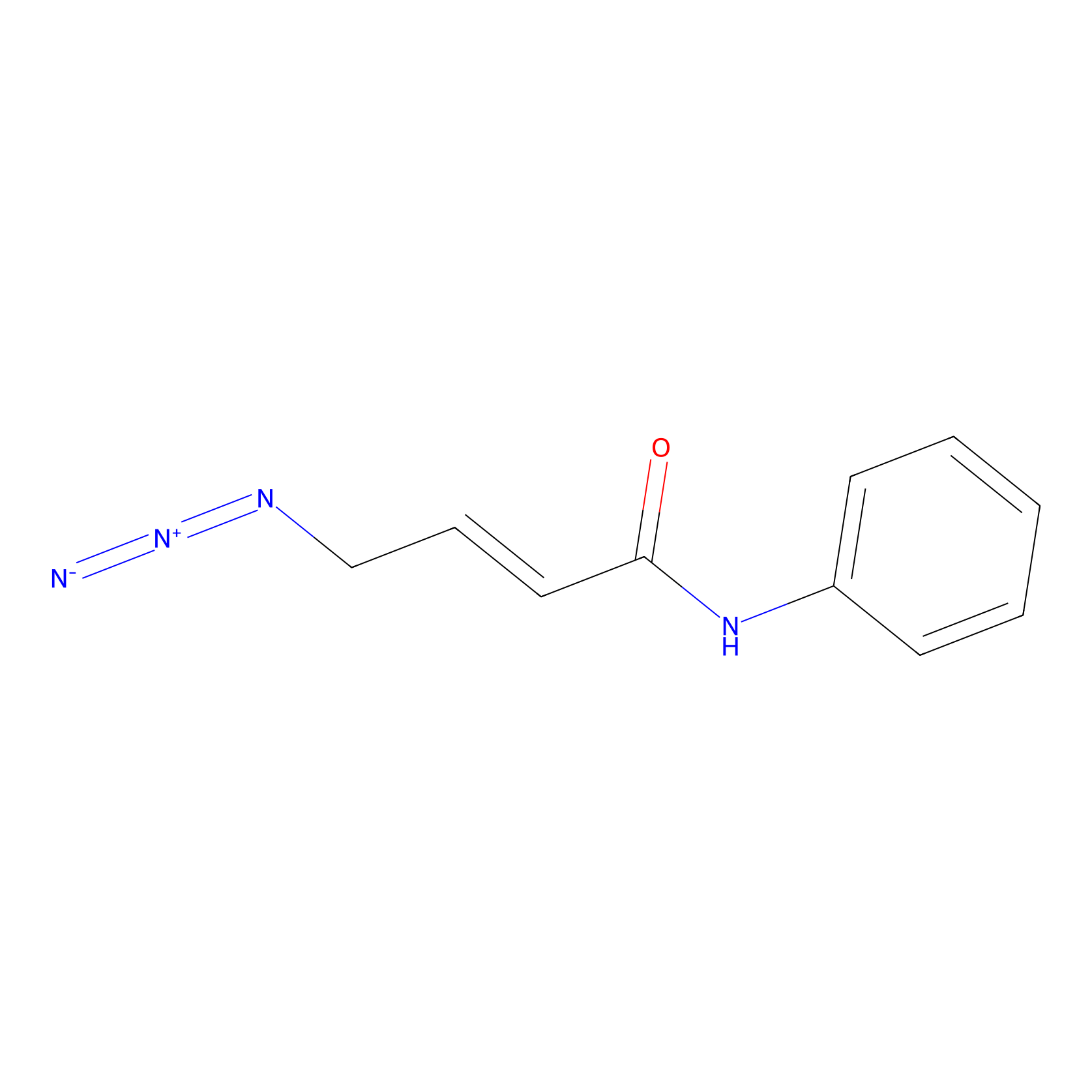
 |
4.02 | LDD0400 | [6] | |
|
DA-P3 Probe Info |
 |
5.98 | LDD0179 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0167 | [8] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 13.14 | LDD0403 | [1] |
| LDCM0027 | Dopamine | HEK-293T | 5.98 | LDD0179 | [7] |
| LDCM0022 | KB02 | A2058 | C283(1.75) | LDD2253 | [5] |
| LDCM0023 | KB03 | A2058 | C283(2.00) | LDD2670 | [5] |
| LDCM0024 | KB05 | COLO792 | C283(2.03) | LDD3310 | [5] |
| LDCM0030 | Luteolin | HEK-293T | 4.90 | LDD0182 | [7] |
| LDCM0154 | YY4 | T cell | 4.02 | LDD0400 | [6] |
The Interaction Atlas With This Target
References
