Details of the Target
General Information of Target
| Target ID | LDTP14192 | |||||
|---|---|---|---|---|---|---|
| Target Name | MORF4 family-associated protein 1 (MRFAP1) | |||||
| Gene Name | MRFAP1 | |||||
| Gene ID | 93621 | |||||
| Synonyms |
PAM14; PGR1; MORF4 family-associated protein 1; Protein PGR1; Protein associated with MRG of 14 kDa |
|||||
| 3D Structure | ||||||
| Sequence |
MQEGELAISPISPVAAMPPLGTHVQARCEAQINLLGEGGICKLPGRLRIQPALWSREDVL
HWLRWAEQEYSLPCTAEHGFEMNGRALCILTKDDFRHRAPSSGDVLYELLQYIKTQRRAL VCGPFFGGIFRLKTPTQHSPVPPEEVTGPSQMDTRRGHLLQPPDPGLTSNFGHLDDPGLA RWTPGKEESLNLCHCAELGCRTQGVCSFPAMPQAPIDGRIADCRLLWDYVYQLLLDTRYE PYIKWEDKDAKIFRVVDPNGLARLWGNHKNRVNMTYEKMSRALRHYYKLNIIKKEPGQKL LFRFLKTPGKMVQDKHSHLEPLESQEQDRIEFKDKRPEISP |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
MORF4 family-associated protein family
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
13.63 | LDD0403 | [1] | |
|
DA-P3 Probe Info |
 |
4.13 | LDD0182 | [2] | |
|
IA-alkyne Probe Info |
 |
C97(0.75) | LDD2182 | [3] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0224 | [4] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2225 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [6] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [7] | |
|
AOyne Probe Info |
 |
13.90 | LDD0443 | [8] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C052 Probe Info |
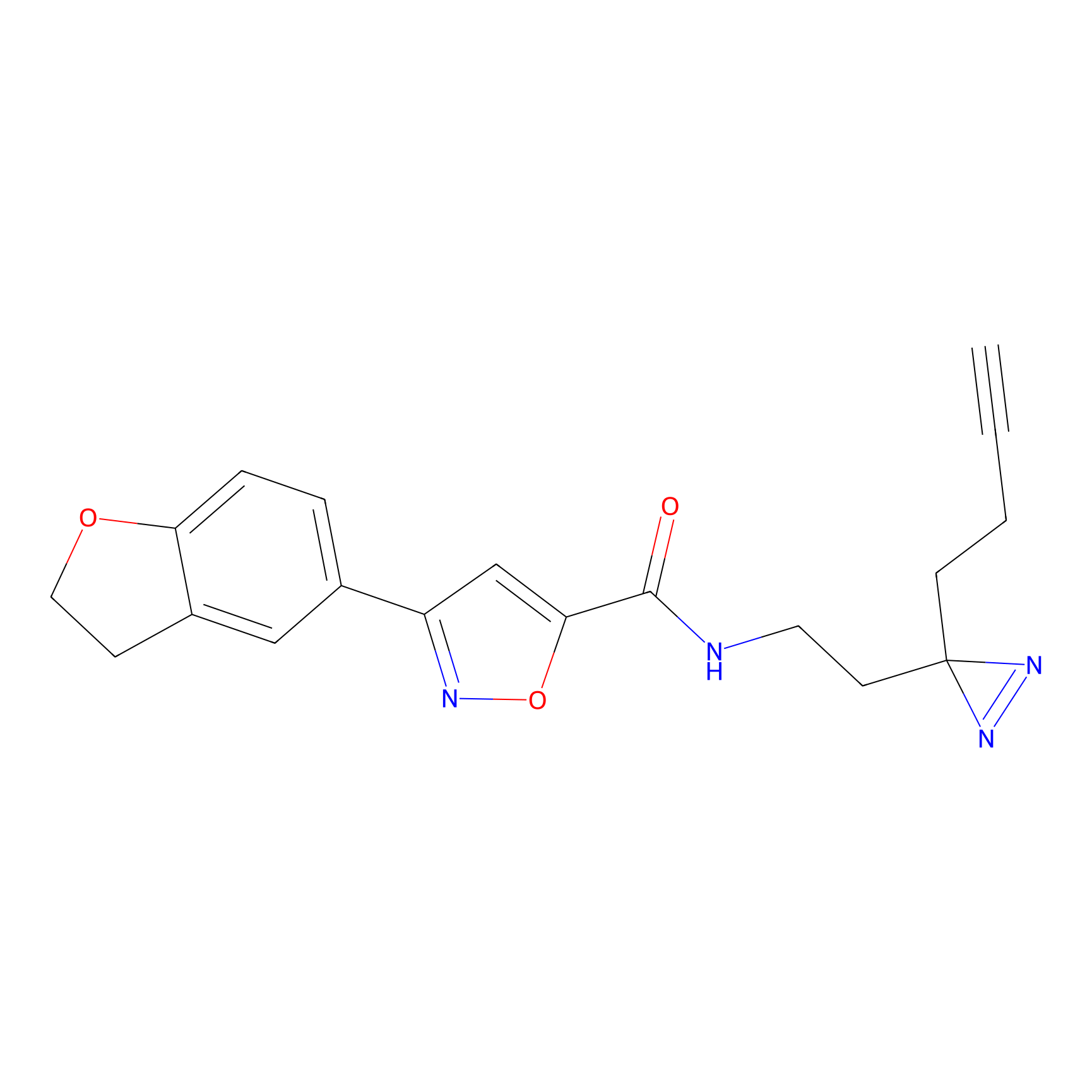 |
6.23 | LDD1750 | [9] | |
|
C299 Probe Info |
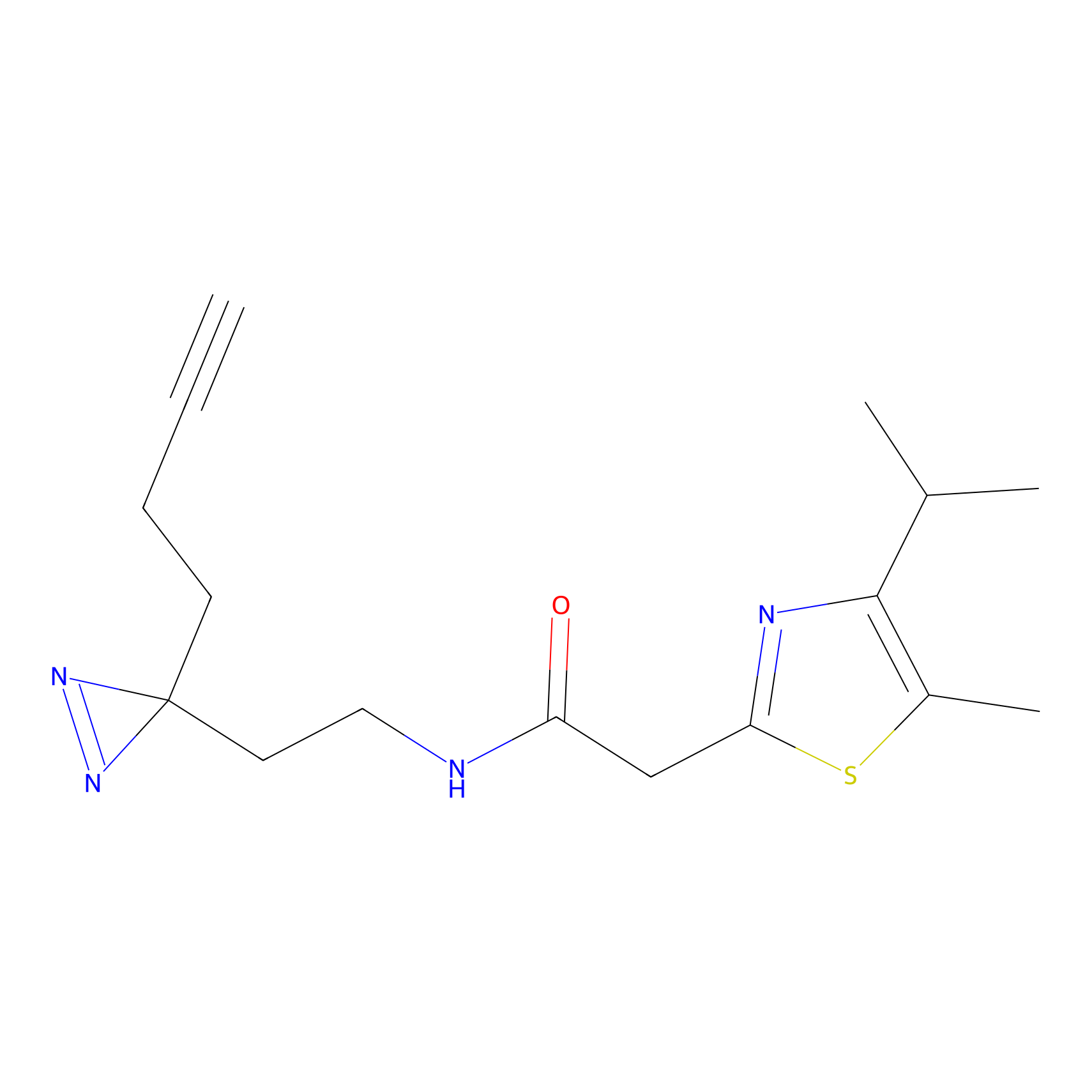 |
8.82 | LDD1968 | [9] | |
|
C305 Probe Info |
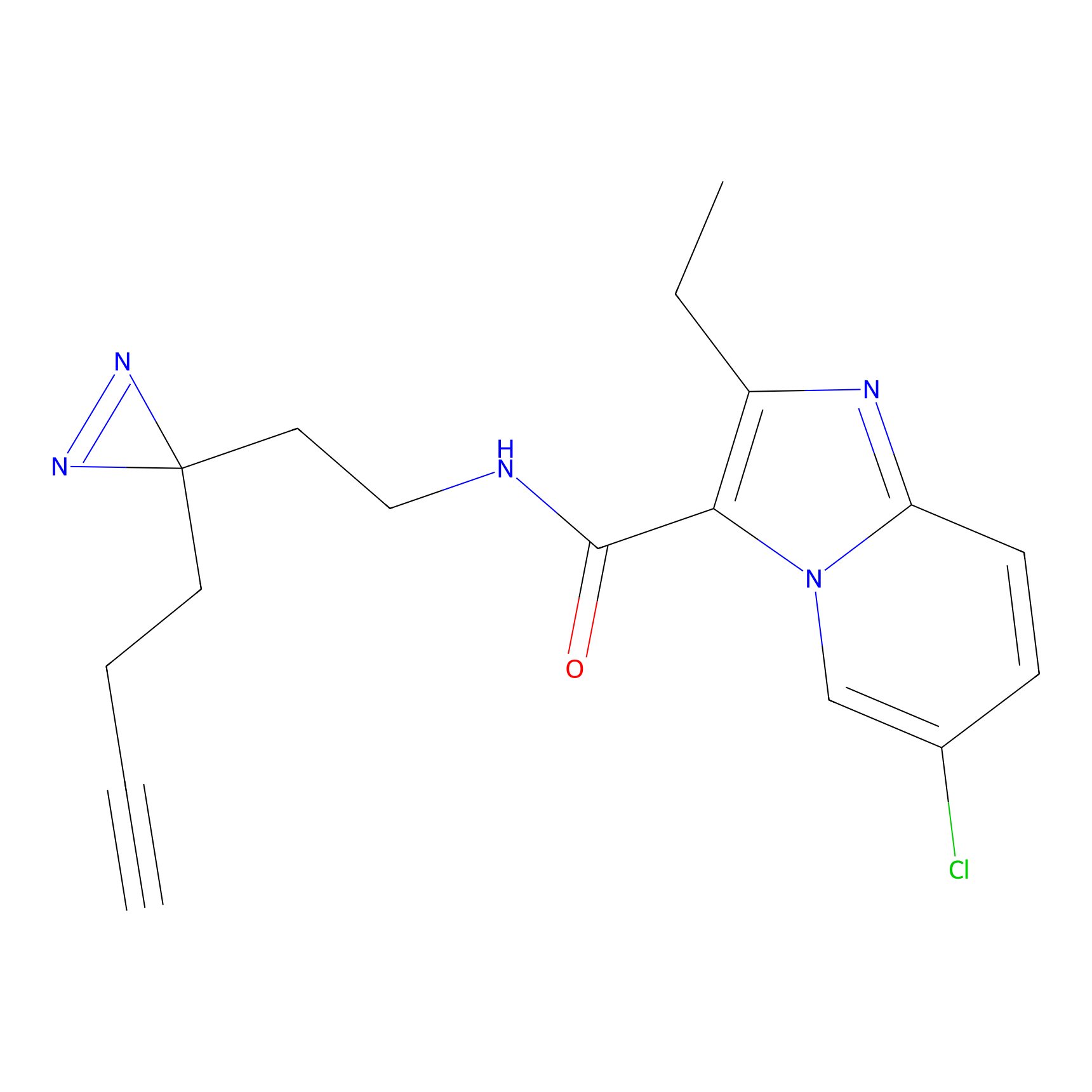 |
9.78 | LDD1974 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0156 | Aniline | NCI-H1299 | 13.63 | LDD0403 | [1] |
| LDCM0632 | CL-Sc | Hep-G2 | C97(1.31) | LDD2227 | [5] |
| LDCM0625 | F8 | Ramos | C97(1.00) | LDD2187 | [3] |
| LDCM0572 | Fragment10 | Ramos | C97(0.12) | LDD2189 | [3] |
| LDCM0573 | Fragment11 | Ramos | C97(20.00) | LDD2190 | [3] |
| LDCM0574 | Fragment12 | Ramos | C97(0.10) | LDD2191 | [3] |
| LDCM0575 | Fragment13 | Ramos | C97(0.13) | LDD2192 | [3] |
| LDCM0576 | Fragment14 | Ramos | C97(0.18) | LDD2193 | [3] |
| LDCM0579 | Fragment20 | Ramos | C97(0.19) | LDD2194 | [3] |
| LDCM0580 | Fragment21 | Ramos | C97(0.17) | LDD2195 | [3] |
| LDCM0582 | Fragment23 | Ramos | C97(0.79) | LDD2196 | [3] |
| LDCM0578 | Fragment27 | Ramos | C97(0.17) | LDD2197 | [3] |
| LDCM0586 | Fragment28 | Ramos | C97(0.84) | LDD2198 | [3] |
| LDCM0588 | Fragment30 | Ramos | C97(0.13) | LDD2199 | [3] |
| LDCM0589 | Fragment31 | Ramos | C97(0.37) | LDD2200 | [3] |
| LDCM0590 | Fragment32 | Ramos | C97(0.16) | LDD2201 | [3] |
| LDCM0468 | Fragment33 | Ramos | C97(0.26) | LDD2202 | [3] |
| LDCM0596 | Fragment38 | Ramos | C97(0.84) | LDD2203 | [3] |
| LDCM0566 | Fragment4 | Ramos | C97(0.75) | LDD2184 | [3] |
| LDCM0610 | Fragment52 | Ramos | C97(0.25) | LDD2204 | [3] |
| LDCM0614 | Fragment56 | Ramos | C97(0.36) | LDD2205 | [3] |
| LDCM0569 | Fragment7 | Ramos | C97(0.56) | LDD2186 | [3] |
| LDCM0571 | Fragment9 | Ramos | C97(0.20) | LDD2188 | [3] |
| LDCM0022 | KB02 | Ramos | C97(0.75) | LDD2182 | [3] |
| LDCM0023 | KB03 | Ramos | C97(1.12) | LDD2183 | [3] |
| LDCM0024 | KB05 | Ramos | C97(1.29) | LDD2185 | [3] |
| LDCM0030 | Luteolin | HEK-293T | 4.13 | LDD0182 | [2] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Huntingtin (HTT) | Huntingtin family | P42858 | |||
| Syntenin-1 (SDCBP) | . | O00560 | |||
| Wolframin (WFS1) | . | O76024 | |||
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger protein 655 (ZNF655) | Krueppel C2H2-type zinc-finger protein family | Q8N720 | |||
| Krueppel-like factor 11 (KLF11) | Sp1 C2H2-type zinc-finger protein family | O14901 | |||
Other
References
