Details of the Target
General Information of Target
| Target ID | LDTP13255 | |||||
|---|---|---|---|---|---|---|
| Target Name | Max-like protein X (MLX) | |||||
| Gene Name | MLX | |||||
| Gene ID | 6945 | |||||
| Synonyms |
BHLHD13; TCFL4; Max-like protein X; Class D basic helix-loop-helix protein 13; bHLHd13; Max-like bHLHZip protein; Protein BigMax; Transcription factor-like protein 4 |
|||||
| 3D Structure | ||||||
| Sequence |
MEGESVKLSSQTLIQAGDDEKNQRTITVNPAHMGKAFKVMNELRSKQLLCDVMIVAEDVE
IEAHRVVLAACSPYFCAMFTGDMSESKAKKIEIKDVDGQTLSKLIDYIYTAEIEVTEENV QVLLPAASLLQLMDVRQNCCDFLQSQLHPTNCLGIRAFADVHTCTDLLQQANAYAEQHFP EVMLGEEFLSLSLDQVCSLISSDKLTVSSEEKVFEAVISWINYEKETRLEHMAKLMEHVR LPLLPRDYLVQTVEEEALIKNNNTCKDFLIEAMKYHLLPLDQRLLIKNPRTKPRTPVSLP KVMIVVGGQAPKAIRSVECYDFEEDRWDQIAELPSRRCRAGVVFMAGHVYAVGGFNGSLR VRTVDVYDGVKDQWTSIASMQERRSTLGAAVLNDLLYAVGGFDGSTGLASVEAYSYKTNE WFFVAPMNTRRSSVGVGVVEGKLYAVGGYDGASRQCLSTVEQYNPATNEWIYVADMSTRR SGAGVGVLSGQLYATGGHDGPLVRKSVEVYDPGTNTWKQVADMNMCRRNAGVCAVNGLLY VVGGDDGSCNLASVEYYNPVTDKWTLLPTNMSTGRSYAGVAVIHKSL |
|||||
| Target Bioclass |
Transcription factor
|
|||||
| Subcellular location |
Cytoplasm; Nucleus
|
|||||
| Function |
Transcription regulator. Forms a sequence-specific DNA-binding protein complex with MAD1, MAD4, MNT, WBSCR14 and MLXIP which recognizes the core sequence 5'-CACGTG-3'. The TCFL4-MAD1, TCFL4-MAD4, TCFL4-WBSCR14 complexes are transcriptional repressors. Plays a role in transcriptional activation of glycolytic target genes. Involved in glucose-responsive gene regulation.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
JZ128-DTB Probe Info |
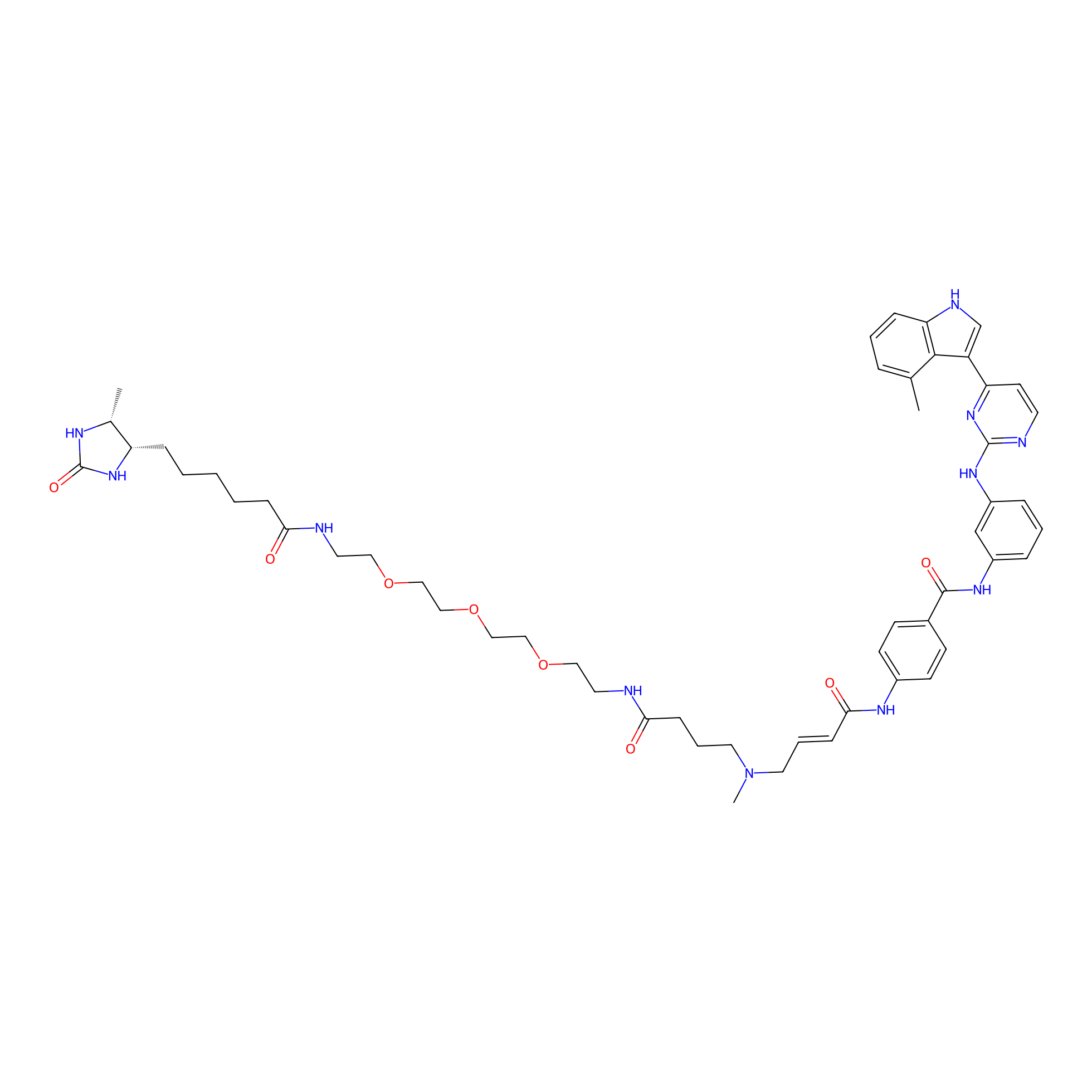 |
N.A. | LDD0462 | [1] | |
|
IA-alkyne Probe Info |
 |
C159(6.28) | LDD1709 | [2] | |
|
DBIA Probe Info |
 |
C159(1.20) | LDD1514 | [3] | |
|
IPM Probe Info |
 |
N.A. | LDD2156 | [4] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [5] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [6] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [7] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0284 | AC24 | HEK-293T | C159(0.98) | LDD1523 | [3] |
| LDCM0293 | AC32 | HEK-293T | C159(1.15) | LDD1532 | [3] |
| LDCM0302 | AC40 | HEK-293T | C159(1.28) | LDD1541 | [3] |
| LDCM0310 | AC48 | HEK-293T | C159(1.01) | LDD1549 | [3] |
| LDCM0319 | AC56 | HEK-293T | C159(1.08) | LDD1558 | [3] |
| LDCM0328 | AC64 | HEK-293T | C159(0.97) | LDD1567 | [3] |
| LDCM0345 | AC8 | HEK-293T | C159(1.30) | LDD1569 | [3] |
| LDCM0275 | AKOS034007705 | HEK-293T | C159(1.20) | LDD1514 | [3] |
| LDCM0369 | CL100 | HEK-293T | C159(0.93) | LDD1573 | [3] |
| LDCM0373 | CL104 | HEK-293T | C159(1.25) | LDD1577 | [3] |
| LDCM0377 | CL108 | HEK-293T | C159(0.90) | LDD1581 | [3] |
| LDCM0382 | CL112 | HEK-293T | C159(1.13) | LDD1586 | [3] |
| LDCM0386 | CL116 | HEK-293T | C159(1.10) | LDD1590 | [3] |
| LDCM0390 | CL12 | HEK-293T | C159(1.14) | LDD1594 | [3] |
| LDCM0391 | CL120 | HEK-293T | C159(1.15) | LDD1595 | [3] |
| LDCM0395 | CL124 | HEK-293T | C159(1.13) | LDD1599 | [3] |
| LDCM0399 | CL128 | HEK-293T | C159(1.22) | LDD1603 | [3] |
| LDCM0403 | CL16 | HEK-293T | C159(1.00) | LDD1607 | [3] |
| LDCM0412 | CL24 | HEK-293T | C159(1.20) | LDD1616 | [3] |
| LDCM0416 | CL28 | HEK-293T | C159(1.36) | LDD1620 | [3] |
| LDCM0425 | CL36 | HEK-293T | C159(1.04) | LDD1629 | [3] |
| LDCM0429 | CL4 | HEK-293T | C159(1.13) | LDD1633 | [3] |
| LDCM0430 | CL40 | HEK-293T | C159(1.04) | LDD1634 | [3] |
| LDCM0438 | CL48 | HEK-293T | C159(0.83) | LDD1642 | [3] |
| LDCM0443 | CL52 | HEK-293T | C159(1.23) | LDD1646 | [3] |
| LDCM0452 | CL60 | HEK-293T | C159(1.26) | LDD1655 | [3] |
| LDCM0456 | CL64 | HEK-293T | C159(1.21) | LDD1659 | [3] |
| LDCM0465 | CL72 | HEK-293T | C159(1.44) | LDD1668 | [3] |
| LDCM0469 | CL76 | HEK-293T | C159(1.24) | LDD1672 | [3] |
| LDCM0478 | CL84 | HEK-293T | C159(0.97) | LDD1681 | [3] |
| LDCM0482 | CL88 | HEK-293T | C159(1.32) | LDD1685 | [3] |
| LDCM0491 | CL96 | HEK-293T | C159(1.16) | LDD1694 | [3] |
| LDCM0625 | F8 | Ramos | C159(1.20) | LDD2187 | [8] |
| LDCM0575 | Fragment13 | Ramos | C159(0.73) | LDD2192 | [8] |
| LDCM0580 | Fragment21 | Ramos | C159(0.57) | LDD2195 | [8] |
| LDCM0582 | Fragment23 | Ramos | C159(0.56) | LDD2196 | [8] |
| LDCM0578 | Fragment27 | Ramos | C159(0.46) | LDD2197 | [8] |
| LDCM0586 | Fragment28 | Ramos | C159(2.41) | LDD2198 | [8] |
| LDCM0588 | Fragment30 | Ramos | C159(0.67) | LDD2199 | [8] |
| LDCM0589 | Fragment31 | Ramos | C159(0.80) | LDD2200 | [8] |
| LDCM0468 | Fragment33 | Ramos | C159(1.28) | LDD2202 | [8] |
| LDCM0614 | Fragment56 | Ramos | C159(0.91) | LDD2205 | [8] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [1] |
| LDCM0024 | KB05 | T cell | C159(6.28) | LDD1709 | [2] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C159(0.51) | LDD2207 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| E3 ubiquitin-protein ligase LNX (LNX1) | . | Q8TBB1 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| AP-4 complex accessory subunit Tepsin (TEPSIN) | . | Q96N21 | |||
Transcription factor
Other
References
