Details of the Target
General Information of Target
| Target ID | LDTP11304 | |||||
|---|---|---|---|---|---|---|
| Target Name | Mitochondrial import inner membrane translocase subunit Tim21 (TIMM21) | |||||
| Gene Name | TIMM21 | |||||
| Gene ID | 29090 | |||||
| Synonyms |
C18orf55; TIM21; Mitochondrial import inner membrane translocase subunit Tim21; TIM21-like protein, mitochondrial |
|||||
| 3D Structure | ||||||
| Sequence |
MPVKRLREVVSQNHGDHLVLLKDELPCVPPALSANKRLPVGTGTSLNGTSRGSSDLTSAR
NCYQPLLENPMVSESDFSKDVAVQVLPLDKIEENNKQKANDIFISQYTMGQKDALRTVLK QKAQSMPVFKEVKVHLLEDAGIEKDAVTQETRISPSGIDSATTVAAATAAAIATAAPLIK VQSDLEAKVNSVTELLSKLQETDKHLQRVTEQQTSIQRKQEKLHCHDHEKQMNVFMEQHI RHLEKLQQQQIDIQTHFISAALKTSSFQPVSMPSSRAVEKYSVKPEHPNLGSCNPSLYNT FASKQAPLKEVEDTSFDKQKSPLETPAPRRFAPVPVSRDDELSKRENLLEEKENMEVSCH RGNVRLLEQILNNNDSLTRKSESSNTTSLTRSKIGWTPEKTNRFPSCEELETTKVTMQKS DDVLHDLGQKEKETNSMVQPKESLSMLKLPDLPQNSVKLQTTNTTRSVLKDAEKILRGVQ NNKKVLEENLEAIIRAKDGAAMYSLINALSTNREMSEKIRIRKTVDEWIKTISAEIQDEL SRTDYEQKRFDQKNQRTKKGQNMTKDIRTNTQDKTVNKSVIPRKHSQKQIEEHFRNLPMR GMPASSLQKERKEGLLKATTVIQDEDYMLQVYGKPVYQGHRSTLKKGPYLRFNSPSPKSR PQRPKVIERVKGTKVKSIRTQTDFYATKPKKMDSKMKHSVPVLPHGDQQYLFSPSREMPT FSGTLEGHLIPMAILLGQTQSNSDTMPPAGVIVSKPHPVTVTTSIPPSSRKVETGVKKPN IAIVEMKSEKKDPPQLTVQVLPSVDIDSISNSSADVLSPLSSPKEASLPPVQTWIKTPEI MKVDEEEVKFPGTNFDEIIDVIQEEEKCDEIPDSEPILEFNRSVKADSTKYNGPPFPPVA STFQPTADILDKVIERKETLENSLIQWVEQEIMSRIISGLFPVQQQIAPSISVSVSETSE PLTSDIVEGTSSGALQLFVDAGVPVNSNVIKHFVNEALAETIAVMLGDREAKKQGPVATG VSGDASTNETYLPARVCTPLPTPQPTPPCSPSSPAKECVLVKTPDSSPCDSDHDMAFPVK EICAEKGDDMPAIMLVNTPTVTPTTTPPPAAAVFTPTLSDISIDKLKVSSPELPKPWGDG DLPLEEENPNSPQEELHPRAIVMSVAKDEEPESMDFPAQPPPPEPVPFMPFPAGTKAPSP SQMPGSDSSTLESTLSVTVTETETLDKPISEGEILFSCGQKLAPKILEDIGLYLTNLNDS LSSTLHDAVEMEDDPPSEGQVIRMSHKKFHADAILSFAKQNQESAVSQQAVYHSEDLENS VGELSEGQRPQLTAAAENILMGHSLYMQPPVTNTQSLDQQCDPKPLSRQFDTVSGSIYED SCASHGPMSLGELELEPNSKLVLPTTLLTAQENDVNLPVAAEDFSQYQLKQNQDVKQVEH KPSQSYLRVRNKSDIAPSQQQVSPGDMDRTQIELNPYLTCVFSGGKAVPLSASQMPPAKM SVMLPSVNLEDCSQSLSLSTMQEDMESSGADTF |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
TIM21 family
|
|||||
| Subcellular location |
Mitochondrion membrane
|
|||||
| Function |
Participates in the translocation of transit peptide-containing proteins across the mitochondrial inner membrane. Also required for assembly of mitochondrial respiratory chain complex I and complex IV as component of the MITRAC (mitochondrial translation regulation assembly intermediate of cytochrome c oxidase complex) complex. Probably shuttles between the presequence translocase and respiratory-chain assembly intermediates in a process that promotes incorporation of early nuclear-encoded subunits into these complexes.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
C-Sul Probe Info |
 |
2.86 | LDD0066 | [2] | |
|
STPyne Probe Info |
 |
K148(2.56) | LDD0277 | [3] | |
|
BTD Probe Info |
 |
C149(1.84) | LDD1700 | [4] | |
|
EA-probe Probe Info |
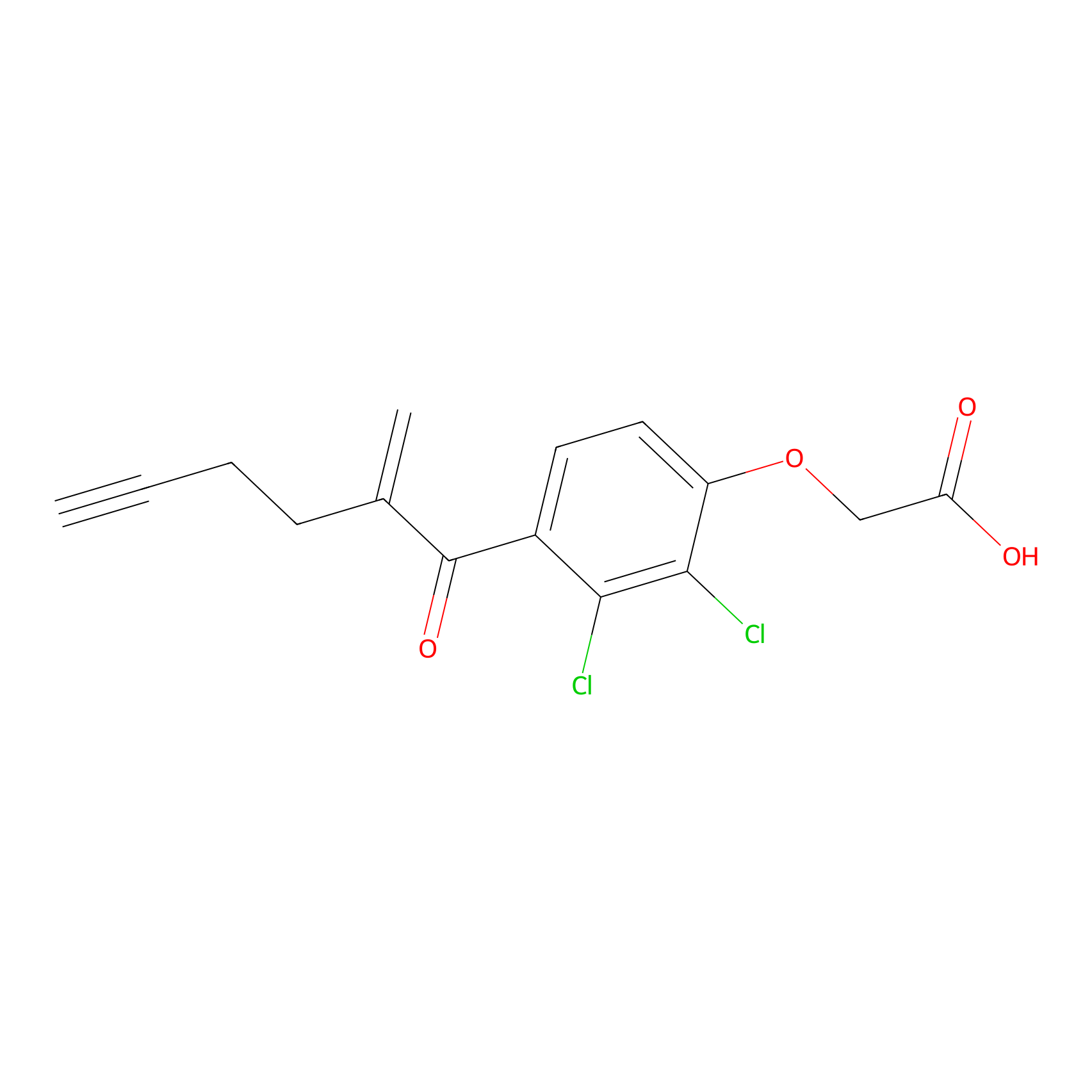 |
N.A. | LDD0440 | [5] | |
|
IA-alkyne Probe Info |
 |
C192(5.68) | LDD1704 | [6] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0223 | [7] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [8] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [8] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [9] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C149(0.84) | LDD2117 | [4] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C149(1.12) | LDD2152 | [4] |
| LDCM0156 | Aniline | NCI-H1299 | 12.30 | LDD0403 | [1] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C149(0.62) | LDD2091 | [4] |
| LDCM0632 | CL-Sc | Hep-G2 | C192(20.00) | LDD2227 | [10] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [5] |
| LDCM0149 | GA | MCF-7 | C192(3.66) | LDD0379 | [11] |
| LDCM0022 | KB02 | T cell-activated | C192(5.68) | LDD1704 | [6] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C149(0.47) | LDD2121 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [7] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C149(0.60) | LDD2089 | [4] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C149(1.03) | LDD2093 | [4] |
| LDCM0504 | Nucleophilic fragment 15a | MDA-MB-231 | C149(1.16) | LDD2097 | [4] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C149(0.91) | LDD2099 | [4] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C149(1.11) | LDD2107 | [4] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C149(0.24) | LDD2108 | [4] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C149(0.57) | LDD2109 | [4] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C149(1.05) | LDD2111 | [4] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C149(3.09) | LDD2119 | [4] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C149(0.71) | LDD2125 | [4] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C149(0.97) | LDD2127 | [4] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C149(1.06) | LDD2129 | [4] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C149(1.16) | LDD2135 | [4] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C149(1.12) | LDD2136 | [4] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C149(0.91) | LDD2137 | [4] |
| LDCM0211 | Nucleophilic fragment 3b | MDA-MB-231 | C149(1.84) | LDD1700 | [4] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C149(0.75) | LDD2140 | [4] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C149(2.60) | LDD2144 | [4] |
| LDCM0556 | Nucleophilic fragment 8a | MDA-MB-231 | C149(0.70) | LDD2150 | [4] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cytochrome c oxidase subunit 1 (MT-CO1) | Heme-copper respiratory oxidase family | P00395 | |||
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Mitochondrial import inner membrane translocase subunit Tim23 (TIMM23) | Tim17/Tim22/Tim23 family | O14925 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Ubiquilin-2 (UBQLN2) | . | Q9UHD9 | |||
References
