Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K143(5.72); K194(8.12); K311(10.00) | LDD0277 | [1] | |
|
m-APA Probe Info |
 |
11.63 | LDD0403 | [2] | |
|
BTD Probe Info |
 |
C277(1.54); C206(0.79) | LDD2140 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C206(3.04) | LDD0169 | [4] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0221 | [5] | |
|
ATP probe Probe Info |
 |
K288(0.00); K194(0.00) | LDD0199 | [6] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [7] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [5] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C056 Probe Info |
 |
26.35 | LDD1753 | [8] | |
|
C391 Probe Info |
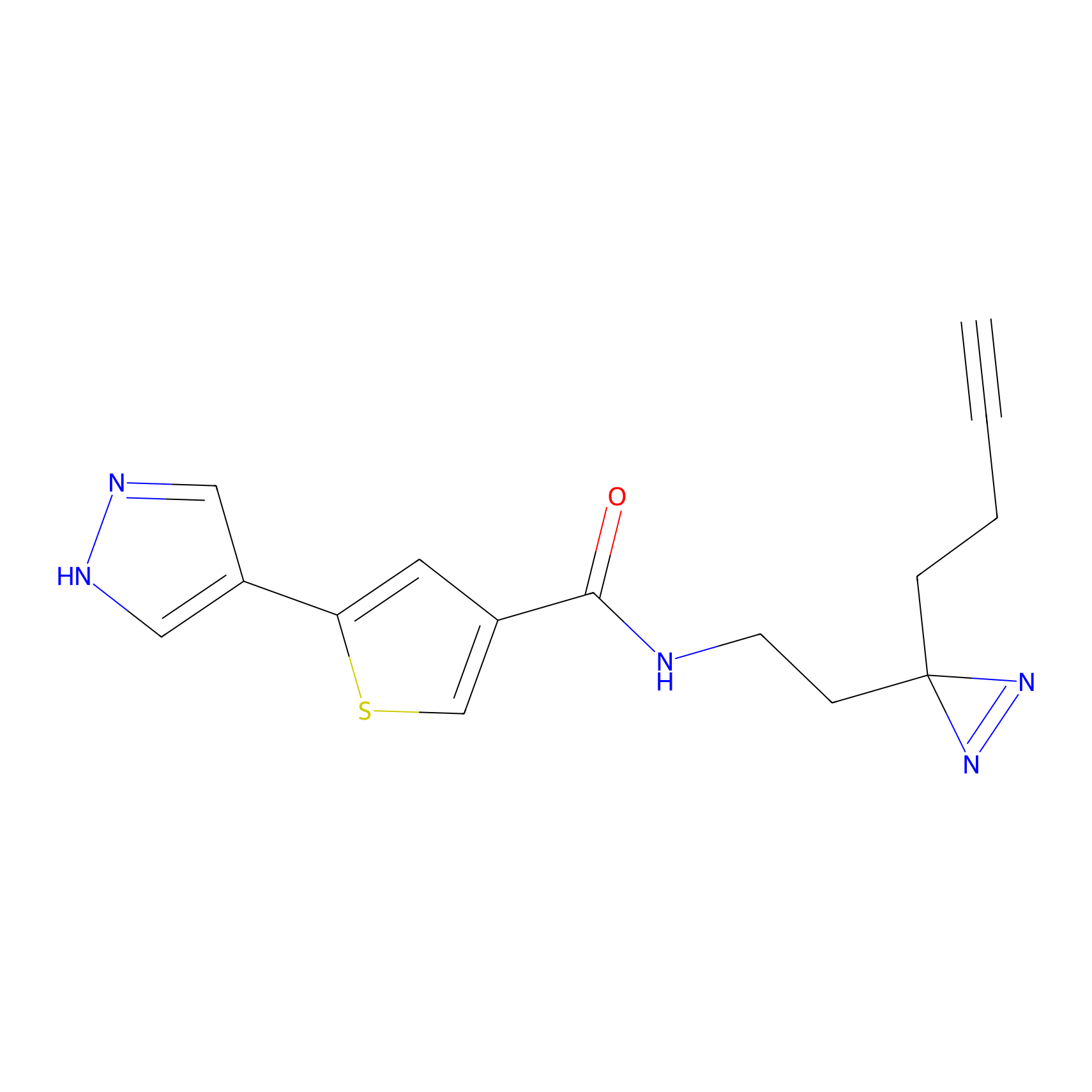 |
41.36 | LDD2050 | [8] | |
|
STS-1 Probe Info |
 |
N.A. | LDD0136 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C206(3.04) | LDD0169 | [4] |
| LDCM0156 | Aniline | NCI-H1299 | 11.63 | LDD0403 | [2] |
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [5] |
| LDCM0625 | F8 | Ramos | C206(1.32) | LDD2187 | [10] |
| LDCM0572 | Fragment10 | Ramos | C206(7.77) | LDD2189 | [10] |
| LDCM0573 | Fragment11 | Ramos | C206(1.09) | LDD2190 | [10] |
| LDCM0575 | Fragment13 | Ramos | C206(1.04) | LDD2192 | [10] |
| LDCM0576 | Fragment14 | Ramos | C206(1.82) | LDD2193 | [10] |
| LDCM0579 | Fragment20 | Ramos | C206(1.87) | LDD2194 | [10] |
| LDCM0580 | Fragment21 | Ramos | C206(1.57) | LDD2195 | [10] |
| LDCM0582 | Fragment23 | Ramos | C206(0.54) | LDD2196 | [10] |
| LDCM0578 | Fragment27 | Ramos | C206(0.80) | LDD2197 | [10] |
| LDCM0586 | Fragment28 | Ramos | C206(0.68) | LDD2198 | [10] |
| LDCM0588 | Fragment30 | Ramos | C206(1.24) | LDD2199 | [10] |
| LDCM0589 | Fragment31 | Ramos | C206(0.63) | LDD2200 | [10] |
| LDCM0468 | Fragment33 | Ramos | C206(2.60) | LDD2202 | [10] |
| LDCM0596 | Fragment38 | Ramos | C206(1.21) | LDD2203 | [10] |
| LDCM0566 | Fragment4 | Ramos | C206(3.25) | LDD2184 | [10] |
| LDCM0610 | Fragment52 | Ramos | C206(1.42) | LDD2204 | [10] |
| LDCM0614 | Fragment56 | Ramos | C206(0.69) | LDD2205 | [10] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [5] |
| LDCM0023 | KB03 | Ramos | C206(3.41) | LDD2183 | [10] |
| LDCM0024 | KB05 | Ramos | C206(2.73) | LDD2185 | [10] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [5] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C277(1.54); C206(0.79) | LDD2140 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Zinc finger and BTB domain-containing protein 14 (ZBTB14) | Krueppel C2H2-type zinc-finger protein family | O43829 | |||
Other
References
