Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K133(4.94) | LDD0277 | [1] | |
|
JZ128-DTB Probe Info |
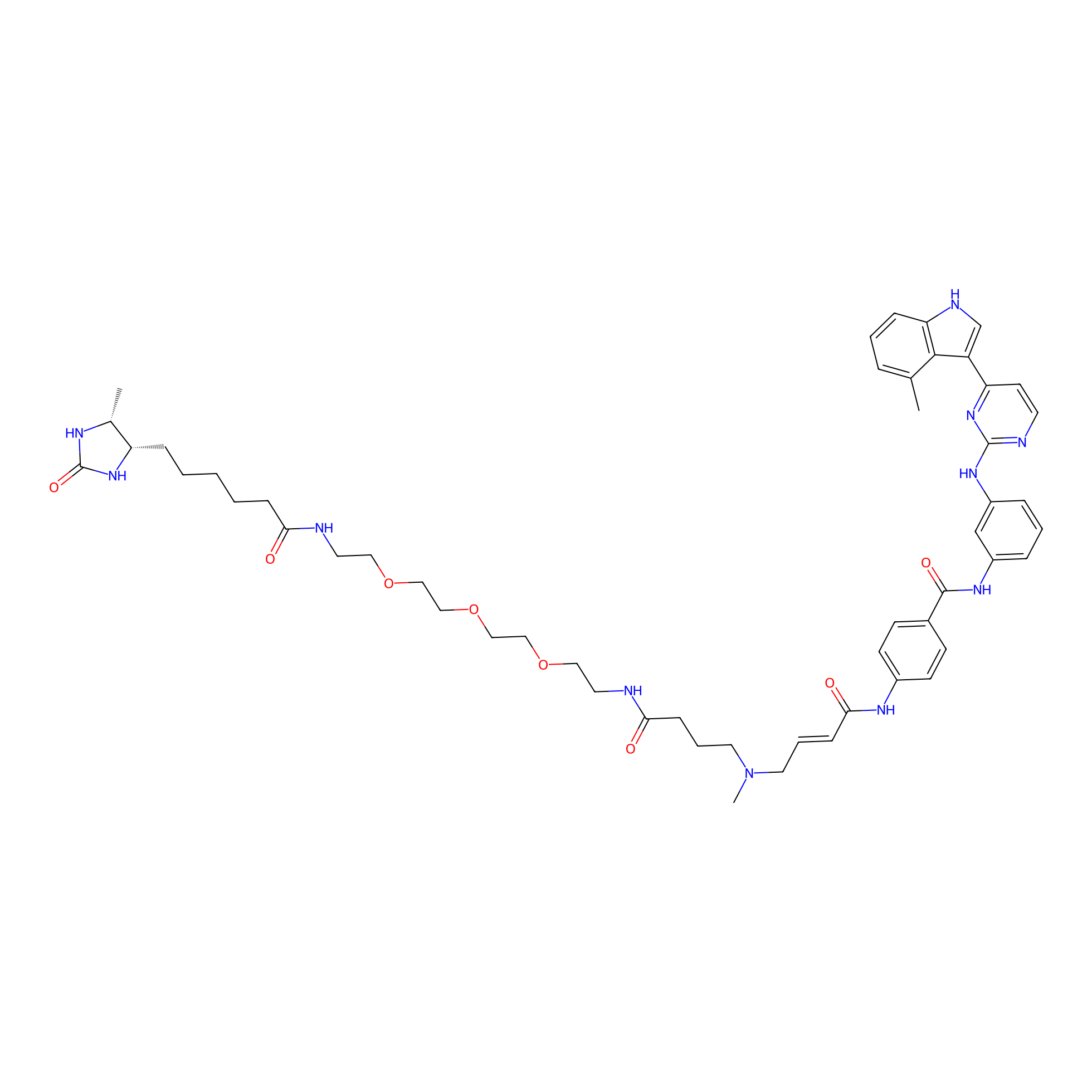 |
N.A. | LDD0462 | [2] | |
|
THZ1-DTB Probe Info |
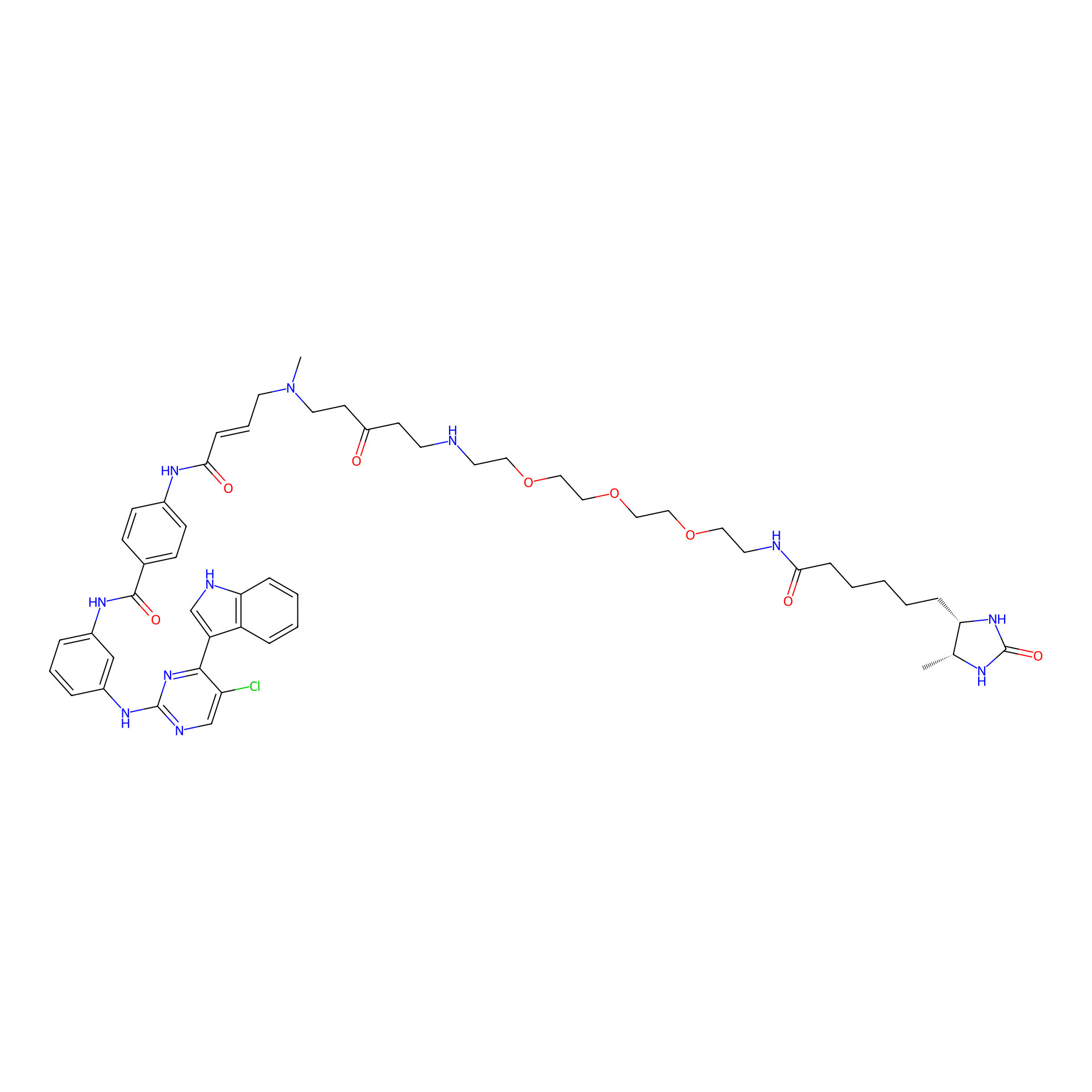 |
C85(1.03) | LDD0460 | [2] | |
|
BTD Probe Info |
 |
C85(1.86) | LDD2090 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C85(11.17) | LDD0170 | [4] | |
|
DBIA Probe Info |
 |
C111(0.96) | LDD1492 | [5] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [6] | |
|
IA-alkyne Probe Info |
 |
C107(0.00); C85(0.00) | LDD0165 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [6] | |
|
IPM Probe Info |
 |
N.A. | LDD0025 | [8] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [8] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C85(1.02) | LDD2117 | [3] |
| LDCM0025 | 4SU-RNA | DM93 | C85(11.17) | LDD0170 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | DM93 | C85(2.91) | LDD0171 | [4] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [2] |
| LDCM0022 | KB02 | HEK-293T | C111(0.96) | LDD1492 | [5] |
| LDCM0023 | KB03 | HEK-293T | C111(1.02) | LDD1497 | [5] |
| LDCM0024 | KB05 | HEK-293T | C111(1.11) | LDD1502 | [5] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C85(1.50) | LDD2102 | [3] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C85(1.86) | LDD2090 | [3] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C85(1.18) | LDD2092 | [3] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C85(1.08) | LDD2098 | [3] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C85(0.89) | LDD2125 | [3] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C85(1.18) | LDD2136 | [3] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C85(0.80) | LDD2137 | [3] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C85(3.28) | LDD2153 | [3] |
| LDCM0021 | THZ1 | HeLa S3 | C85(1.03) | LDD0460 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| RING finger protein 11 (RNF11) | . | Q9Y3C5 | |||
References
