Details of the Target
General Information of Target
| Target ID | LDTP04392 | |||||
|---|---|---|---|---|---|---|
| Target Name | Sodium/potassium-transporting ATPase subunit alpha-2 (ATP1A2) | |||||
| Gene Name | ATP1A2 | |||||
| Gene ID | 477 | |||||
| Synonyms |
KIAA0778; Sodium/potassium-transporting ATPase subunit alpha-2; Na(+)/K(+) ATPase alpha-2 subunit; EC 7.2.2.13; Sodium pump subunit alpha-2 |
|||||
| 3D Structure | ||||||
| Sequence |
MGRGAGREYSPAATTAENGGGKKKQKEKELDELKKEVAMDDHKLSLDELGRKYQVDLSKG
LTNQRAQDVLARDGPNALTPPPTTPEWVKFCRQLFGGFSILLWIGAILCFLAYGIQAAME DEPSNDNLYLGVVLAAVVIVTGCFSYYQEAKSSKIMDSFKNMVPQQALVIREGEKMQINA EEVVVGDLVEVKGGDRVPADLRIISSHGCKVDNSSLTGESEPQTRSPEFTHENPLETRNI CFFSTNCVEGTARGIVIATGDRTVMGRIATLASGLEVGRTPIAMEIEHFIQLITGVAVFL GVSFFVLSLILGYSWLEAVIFLIGIIVANVPEGLLATVTVCLTLTAKRMARKNCLVKNLE AVETLGSTSTICSDKTGTLTQNRMTVAHMWFDNQIHEADTTEDQSGATFDKRSPTWTALS RIAGLCNRAVFKAGQENISVSKRDTAGDASESALLKCIELSCGSVRKMRDRNPKVAEIPF NSTNKYQLSIHEREDSPQSHVLVMKGAPERILDRCSTILVQGKEIPLDKEMQDAFQNAYM ELGGLGERVLGFCQLNLPSGKFPRGFKFDTDELNFPTEKLCFVGLMSMIDPPRAAVPDAV GKCRSAGIKVIMVTGDHPITAKAIAKGVGIISEGNETVEDIAARLNIPMSQVNPREAKAC VVHGSDLKDMTSEQLDEILKNHTEIVFARTSPQQKLIIVEGCQRQGAIVAVTGDGVNDSP ALKKADIGIAMGISGSDVSKQAADMILLDDNFASIVTGVEEGRLIFDNLKKSIAYTLTSN IPEITPFLLFIIANIPLPLGTVTILCIDLGTDMVPAISLAYEAAESDIMKRQPRNSQTDK LVNERLISMAYGQIGMIQALGGFFTYFVILAENGFLPSRLLGIRLDWDDRTMNDLEDSYG QEWTYEQRKVVEFTCHTAFFASIVVVQWADLIICKTRRNSVFQQGMKNKILIFGLLEETA LAAFLSYCPGMGVALRMYPLKVTWWFCAFPYSLLIFIYDEVRKLILRRYPGGWVEKETYY |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Cation transport ATPase (P-type) (TC 3.A.3) family, Type IIC subfamily
|
|||||
| Subcellular location |
Membrane
|
|||||
| Function |
This is the catalytic component of the active enzyme, which catalyzes the hydrolysis of ATP coupled with the exchange of sodium and potassium ions across the plasma membrane. This action creates the electrochemical gradient of sodium and potassium, providing the energy for active transport of various nutrients.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C436(3.78) | LDD3310 | [1] | |
|
THZ1-DTB Probe Info |
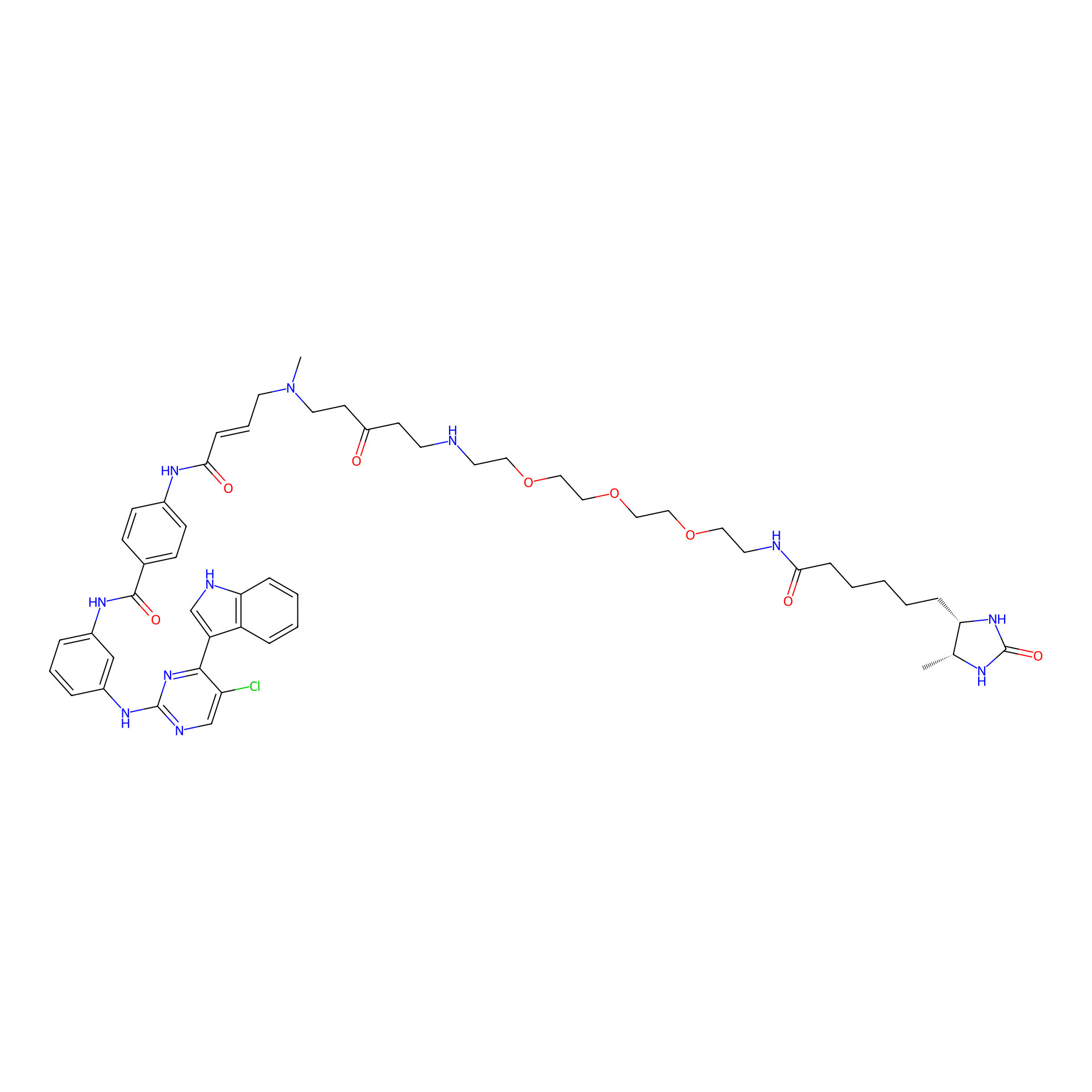 |
C372(1.06) | LDD0460 | [2] | |
|
AHL-Pu-1 Probe Info |
 |
C372(2.27) | LDD0169 | [3] | |
|
IA-alkyne Probe Info |
 |
C372(0.00); C660(0.00) | LDD0032 | [4] | |
|
WYneC Probe Info |
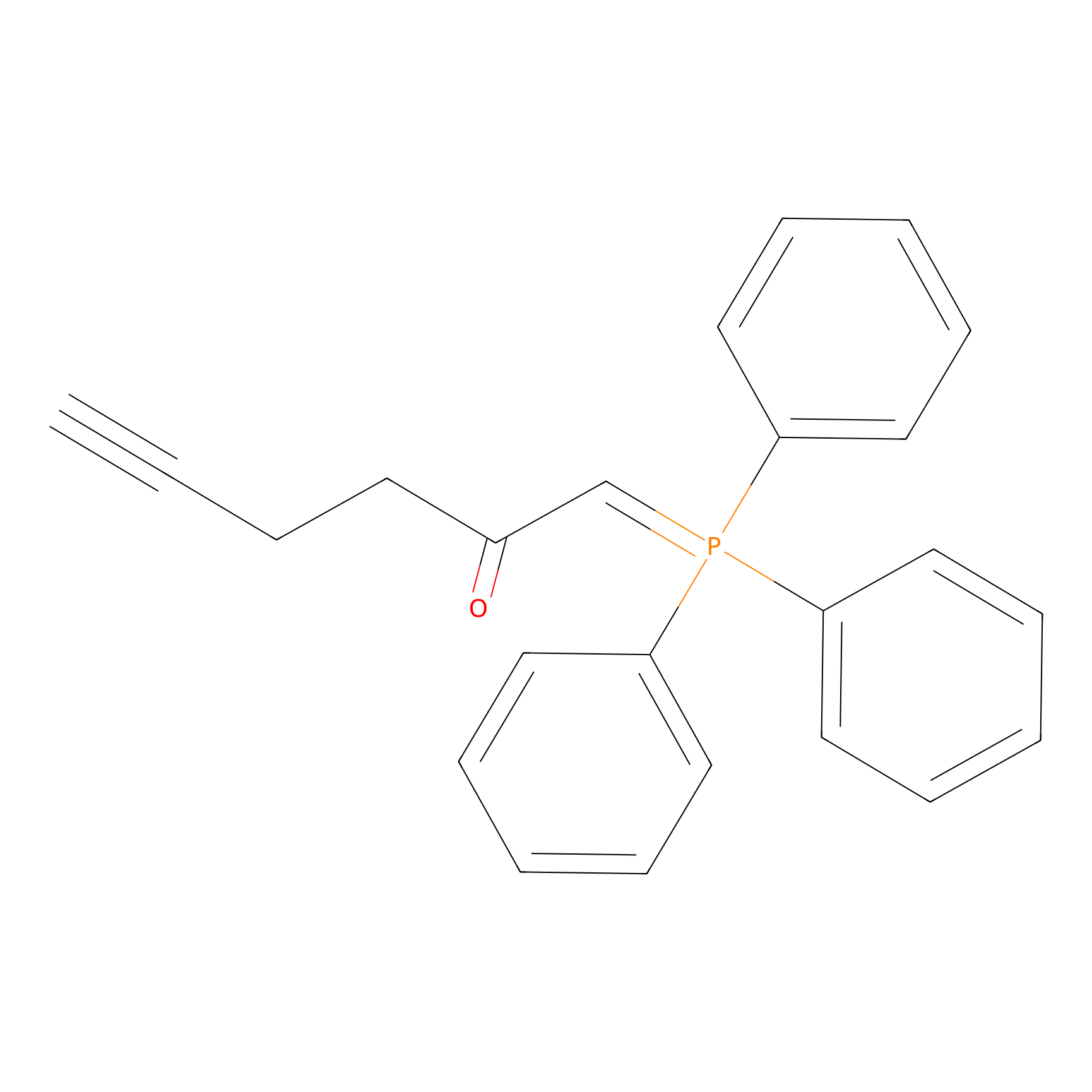 |
N.A. | LDD0014 | [5] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [5] | |
|
Phosphinate-6 Probe Info |
 |
C702(0.00); C372(0.00); C603(0.00) | LDD0018 | [6] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [7] | |
|
Crotonaldehyde Probe Info |
 |
C660(0.00); H663(0.00) | LDD0219 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C372(2.27) | LDD0169 | [3] |
| LDCM0108 | Chloroacetamide | HeLa | C660(0.00); H663(0.00) | LDD0222 | [7] |
| LDCM0107 | IAA | HeLa | C660(0.00); H663(0.00) | LDD0221 | [7] |
| LDCM0022 | KB02 | 22RV1 | C436(2.09); C382(1.41) | LDD2243 | [1] |
| LDCM0023 | KB03 | 22RV1 | C436(2.86); C382(1.99) | LDD2660 | [1] |
| LDCM0024 | KB05 | COLO792 | C436(3.78) | LDD3310 | [1] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [7] |
| LDCM0021 | THZ1 | HeLa S3 | C372(1.06) | LDD0460 | [2] |
The Interaction Atlas With This Target
References
