Details of the Target
General Information of Target
| Target ID | LDTP03891 | |||||
|---|---|---|---|---|---|---|
| Target Name | Nicotinamide N-methyltransferase (NNMT) | |||||
| Gene Name | NNMT | |||||
| Gene ID | 4837 | |||||
| Synonyms |
Nicotinamide N-methyltransferase; EC 2.1.1.1 |
|||||
| 3D Structure | ||||||
| Sequence |
MESGFTSKDTYLSHFNPRDYLEKYYKFGSRHSAESQILKHLLKNLFKIFCLDGVKGDLLI
DIGSGPTIYQLLSACESFKEIVVTDYSDQNLQELEKWLKKEPEAFDWSPVVTYVCDLEGN RVKGPEKEEKLRQAVKQVLKCDVTQSQPLGAVPLPPADCVLSTLCLDAACPDLPTYCRAL RNLGSLLKPGGFLVIMDALKSSYYMIGEQKFSSLPLGREAVEAAVKEAGYTIEWFEVISQ SYSSTMANNEGLFSLVARKLSRPL |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Class I-like SAM-binding methyltransferase superfamily, NNMT/PNMT/TEMT family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Catalyzes the N-methylation of nicotinamide using the universal methyl donor S-adenosyl-L-methionine to form N1-methylnicotinamide and S-adenosyl-L-homocysteine, a predominant nicotinamide/vitamin B3 clearance pathway. Plays a central role in regulating cellular methylation potential, by consuming S-adenosyl-L-methionine and limiting its availability for other methyltransferases. Actively mediates genome-wide epigenetic and transcriptional changes through hypomethylation of repressive chromatin marks, such as H3K27me3. In a developmental context, contributes to low levels of the repressive histone marks that characterize pluripotent embryonic stem cell pre-implantation state. Acts as a metabolic regulator primarily on white adipose tissue energy expenditure as well as hepatic gluconeogenesis and cholesterol biosynthesis. In white adipocytes, regulates polyamine flux by consuming S-adenosyl-L-methionine which provides for propylamine group in polyamine biosynthesis, whereas by consuming nicotinamide controls NAD(+) levels through the salvage pathway. Via its product N1-methylnicotinamide regulates protein acetylation in hepatocytes, by repressing the ubiquitination and increasing the stability of SIRT1 deacetylase. Can also N-methylate other pyridines structurally related to nicotinamide and play a role in xenobiotic detoxification.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K210(5.63); K23(5.21); K26(10.00); K39(10.00) | LDD0277 | [1] | |
|
BTD Probe Info |
 |
C50(3.25) | LDD1699 | [2] | |
|
DBIA Probe Info |
 |
C50(1.48); C115(0.82) | LDD3314 | [3] | |
|
THZ1-DTB Probe Info |
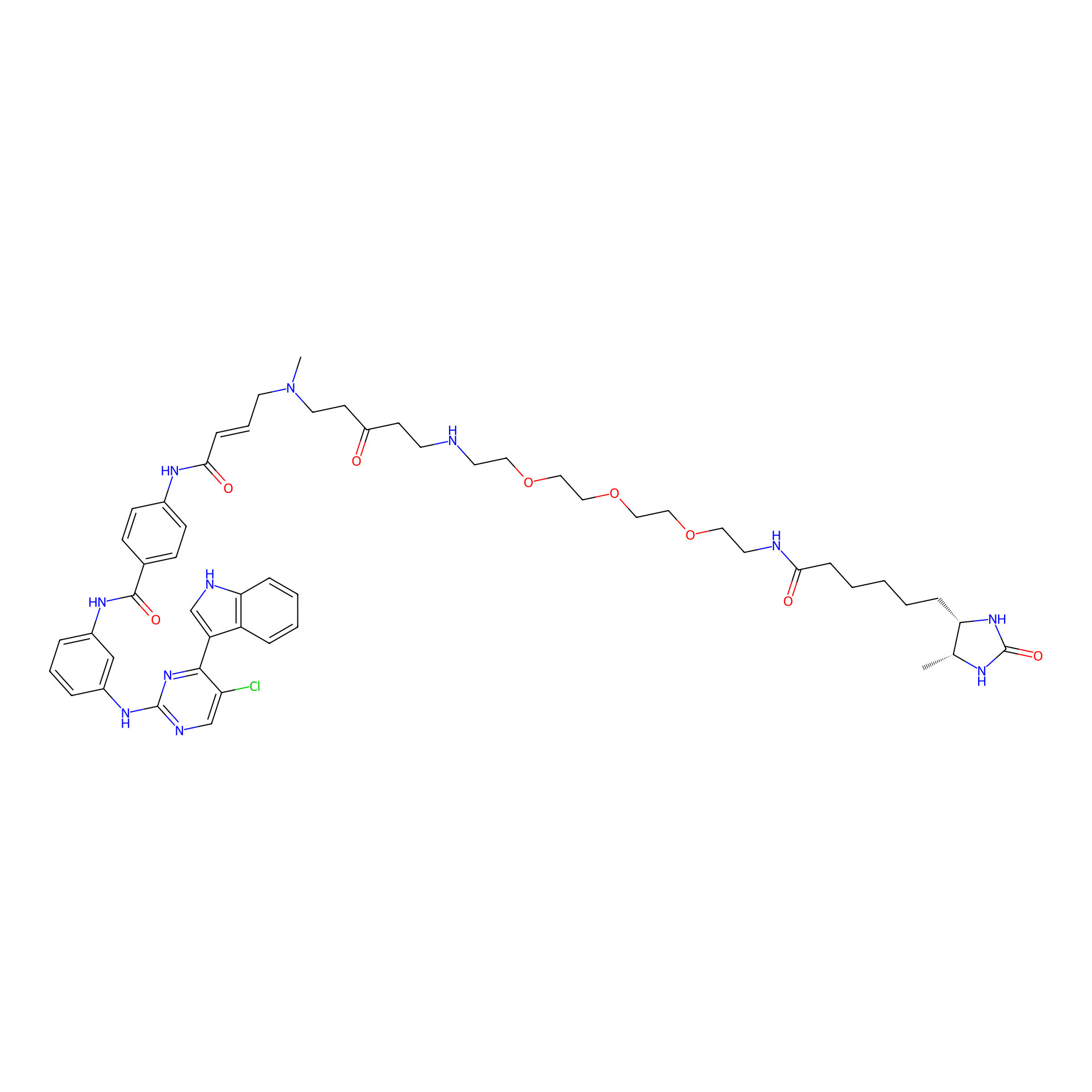 |
C170(1.10); C159(1.05); C165(1.01) | LDD0460 | [4] | |
|
IPM Probe Info |
 |
C50(0.62) | LDD1701 | [2] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [5] | |
|
m-APA Probe Info |
 |
N.A. | LDD2231 | [5] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [6] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0151 | [7] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [8] | |
|
Acrolein Probe Info |
 |
H31(0.00); C50(0.00); H14(0.00) | LDD0217 | [9] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [9] | |
|
HHS-475 Probe Info |
 |
Y20(3.30); Y203(1.77) | LDD2238 | [10] | |
|
HHS-482 Probe Info |
 |
Y11(1.29); Y203(1.05); Y86(1.14) | LDD2239 | [10] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
FFF probe13 Probe Info |
 |
10.88 | LDD0476 | [11] | |
|
FFF probe3 Probe Info |
 |
5.08 | LDD0465 | [11] | |
|
STS-2 Probe Info |
 |
N.A. | LDD0139 | [12] | |
|
DA-2 Probe Info |
 |
N.A. | LDD0072 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C50(0.81) | LDD2117 | [2] |
| LDCM0108 | Chloroacetamide | HeLa | H14(0.00); H31(0.00); C50(0.00) | LDD0222 | [9] |
| LDCM0107 | IAA | HeLa | H14(0.00); H31(0.00); C50(0.00) | LDD0221 | [9] |
| LDCM0022 | KB02 | 42-MG-BA | C75(2.34); C115(2.09) | LDD2244 | [3] |
| LDCM0023 | KB03 | MDA-MB-231 | C50(0.62) | LDD1701 | [2] |
| LDCM0024 | KB05 | IGR37 | C50(1.48); C115(0.82) | LDD3314 | [3] |
| LDCM0109 | NEM | HeLa | H14(0.00); H31(0.00) | LDD0223 | [9] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C50(0.95) | LDD2107 | [2] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C50(0.69) | LDD2109 | [2] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C50(0.96) | LDD2123 | [2] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C50(1.32) | LDD2125 | [2] |
| LDCM0534 | Nucleophilic fragment 30a | MDA-MB-231 | C50(1.46) | LDD2127 | [2] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C50(1.28) | LDD2136 | [2] |
| LDCM0544 | Nucleophilic fragment 39 | MDA-MB-231 | C50(0.85) | LDD2137 | [2] |
| LDCM0021 | THZ1 | HeLa S3 | C170(1.10); C159(1.05); C165(1.01) | LDD0460 | [4] |
The Interaction Atlas With This Target
References
