Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K120(6.22); K128(5.42); K134(9.74); K37(8.04) | LDD0277 | [1] | |
|
AZ-9 Probe Info |
 |
E92(1.09) | LDD2208 | [2] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [1] | |
|
OPA-S-S-alkyne Probe Info |
 |
K37(2.89) | LDD3494 | [3] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0222 | [4] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [5] | |
|
SF Probe Info |
 |
Y40(0.00); K37(0.00); Y143(0.00) | LDD0028 | [6] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [7] | |
|
1d-yne Probe Info |
 |
K119(0.00); K128(0.00); K96(0.00) | LDD0357 | [7] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [4] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [4] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C292 Probe Info |
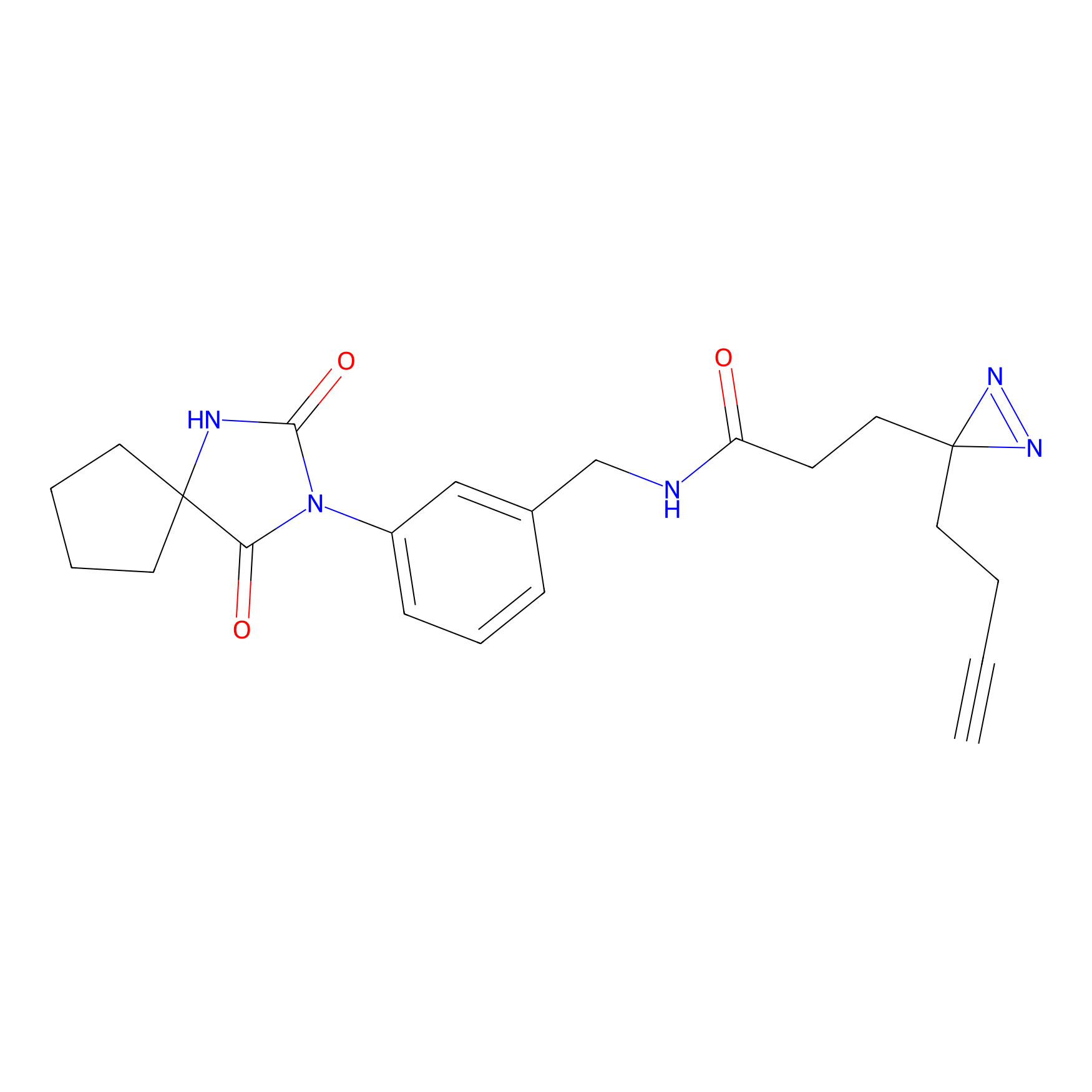 |
5.98 | LDD1962 | [8] | |
|
C361 Probe Info |
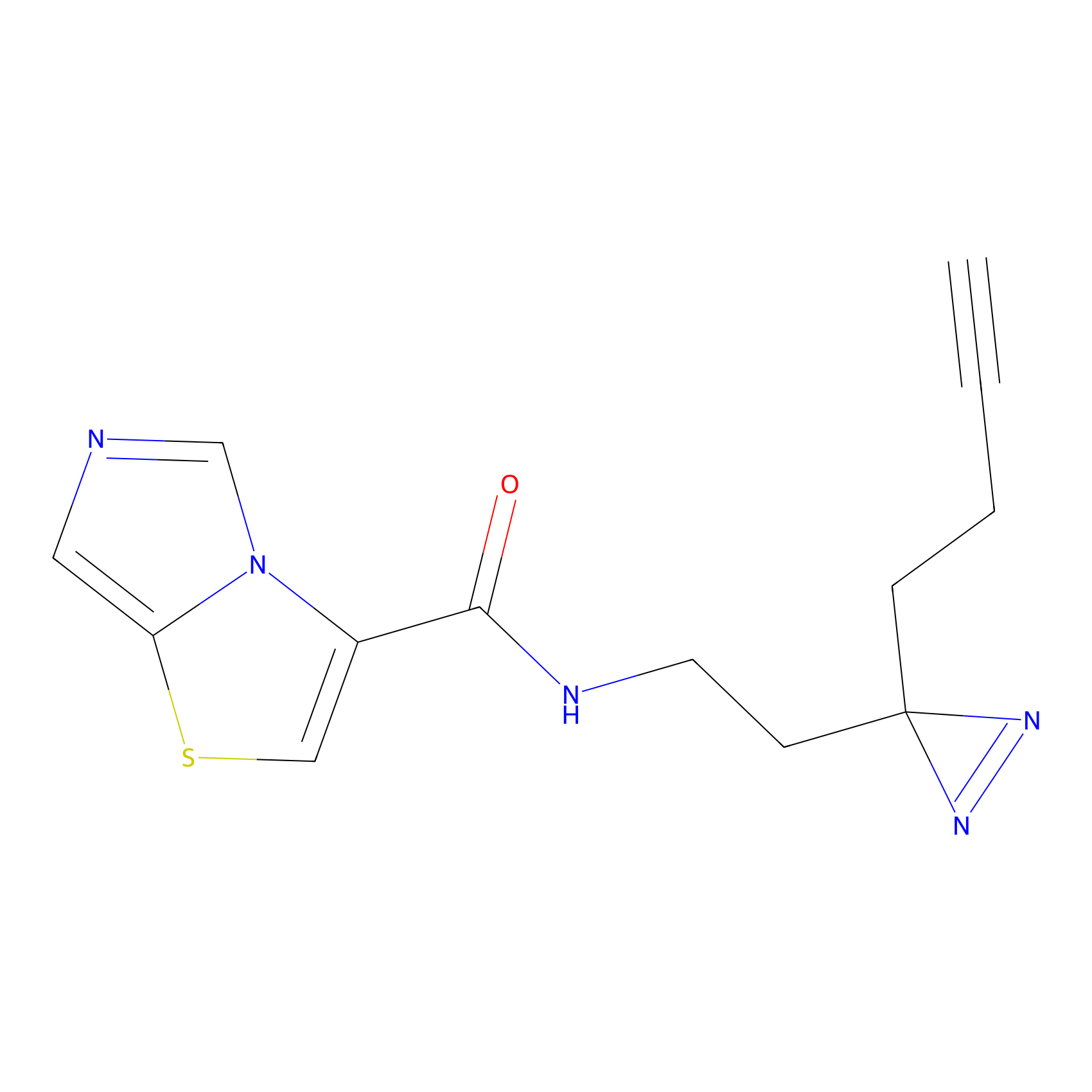 |
18.90 | LDD2022 | [8] | |
|
FFF probe4 Probe Info |
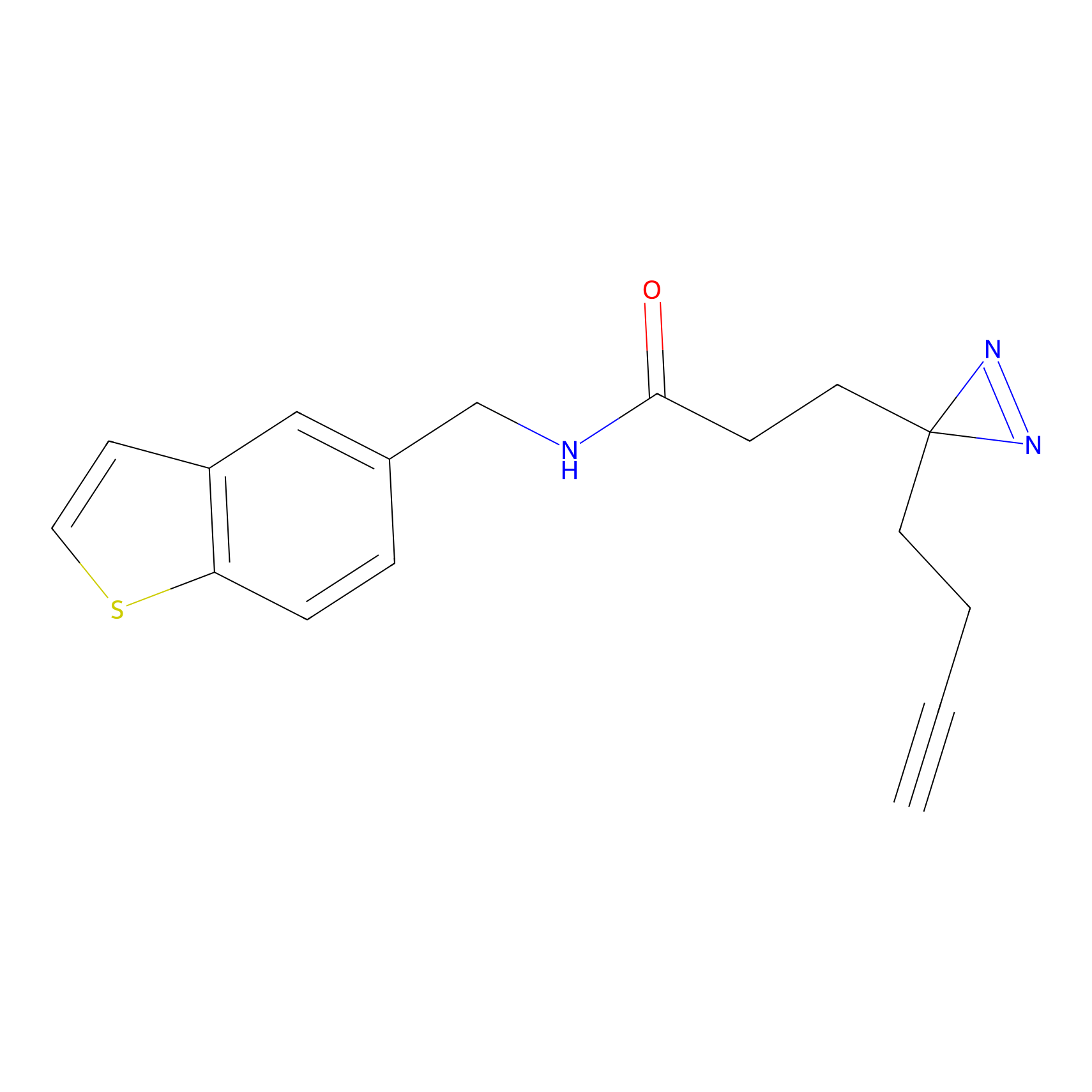 |
5.16 | LDD0466 | [9] | |
Competitor(s) Related to This Target
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
References
