Details of the Target
General Information of Target
| Target ID | LDTP02774 | |||||
|---|---|---|---|---|---|---|
| Target Name | Phosphoglycerate mutase 2 (PGAM2) | |||||
| Gene Name | PGAM2 | |||||
| Gene ID | 5224 | |||||
| Synonyms |
PGAMM; Phosphoglycerate mutase 2; EC 5.4.2.11; EC 5.4.2.4; BPG-dependent PGAM 2; Muscle-specific phosphoglycerate mutase; Phosphoglycerate mutase isozyme M; PGAM-M |
|||||
| 3D Structure | ||||||
| Sequence |
MATHRLVMVRHGESTWNQENRFCGWFDAELSEKGTEEAKRGAKAIKDAKMEFDICYTSVL
KRAIRTLWAILDGTDQMWLPVVRTWRLNERHYGGLTGLNKAETAAKHGEEQVKIWRRSFD IPPPPMDEKHPYYNSISKERRYAGLKPGELPTCESLKDTIARALPFWNEEIVPQIKAGKR VLIAAHGNSLRGIVKHLEGMSDQAIMELNLPTGIPIVYELNKELKPTKPMQFLGDEETVR KAMEAVAAQGKAK |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Phosphoglycerate mutase family, BPG-dependent PGAM subfamily
|
|||||
| Function | Interconversion of 3- and 2-phosphoglycerate with 2,3-bisphosphoglycerate as the primer of the reaction. Can also catalyze the reaction of EC 5.4.2.4 (synthase), but with a reduced activity. | |||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
P3 Probe Info |
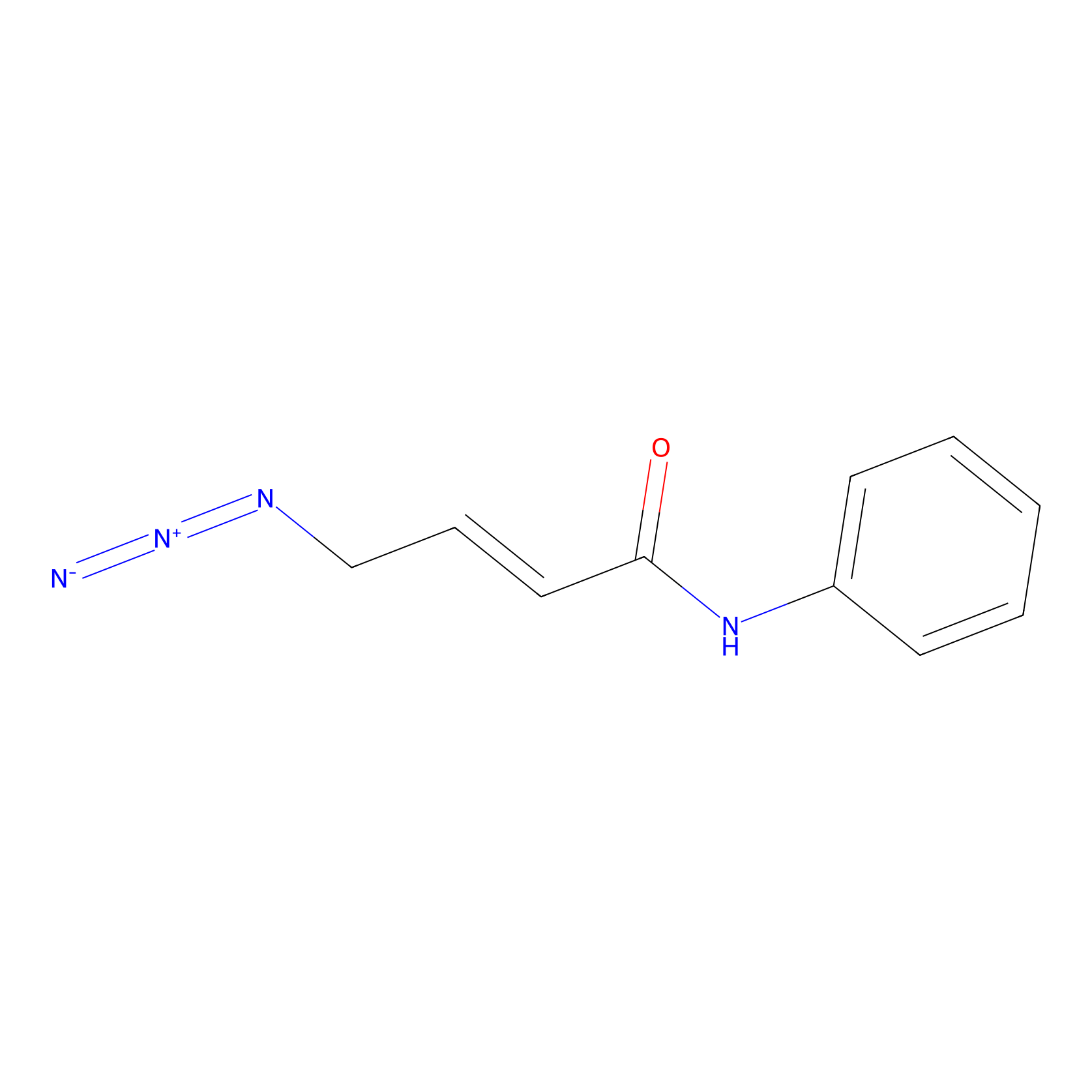
 |
1.54 | LDD0454 | [1] | |
|
P8 Probe Info |
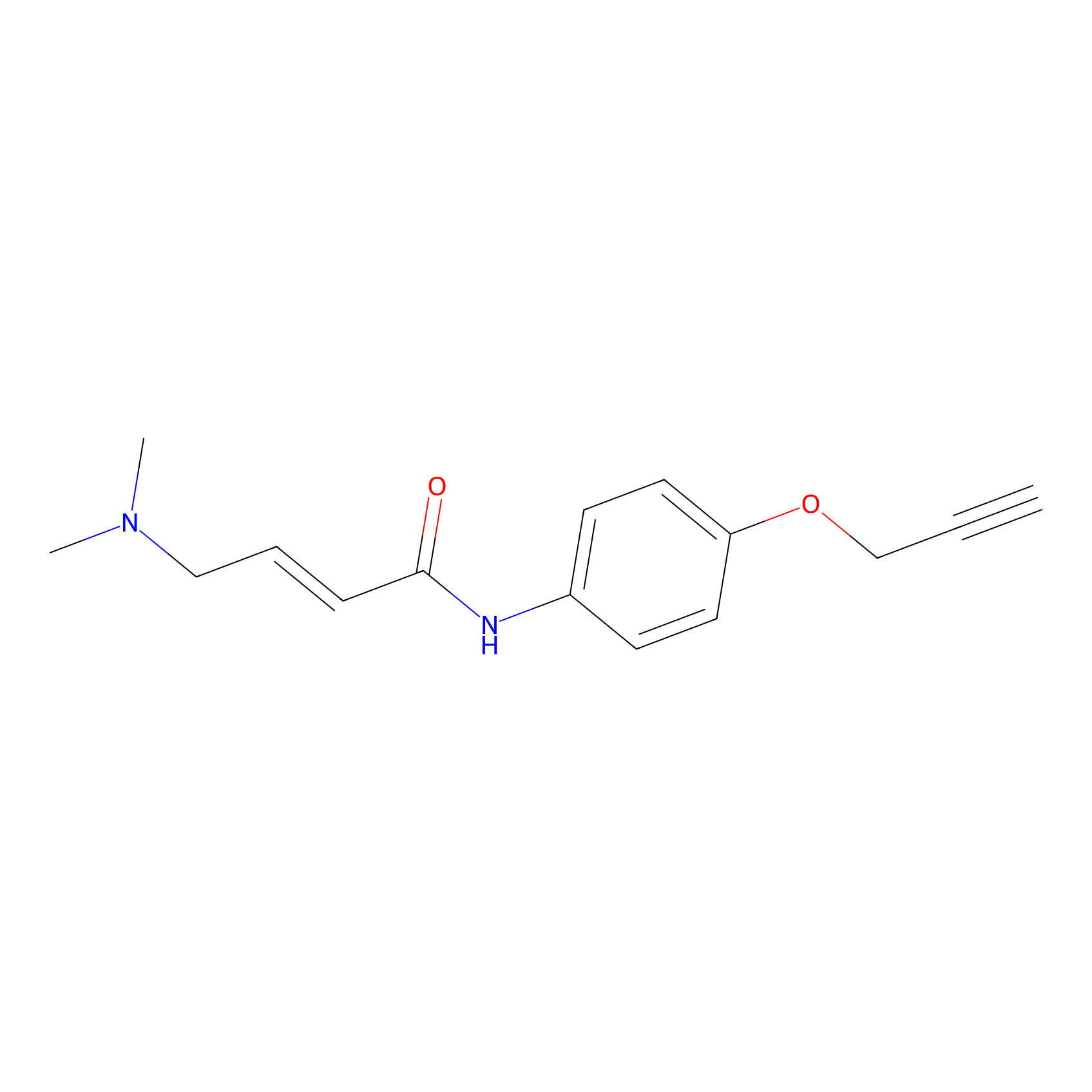
 |
5.31 | LDD0455 | [1] | |
|
TH211 Probe Info |
 |
Y92(15.20) | LDD0260 | [2] | |
|
AZ-9 Probe Info |
 |
E244(0.99) | LDD2208 | [3] | |
|
Probe 1 Probe Info |
 |
Y92(15.15) | LDD3495 | [4] | |
|
DBIA Probe Info |
 |
C55(1.04); C153(1.20) | LDD3377 | [5] | |
|
HHS-475 Probe Info |
 |
Y92(0.97) | LDD0264 | [6] | |
|
HHS-465 Probe Info |
 |
Y92(10.00) | LDD2237 | [7] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [8] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [8] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [8] | |
|
MPP-AC Probe Info |
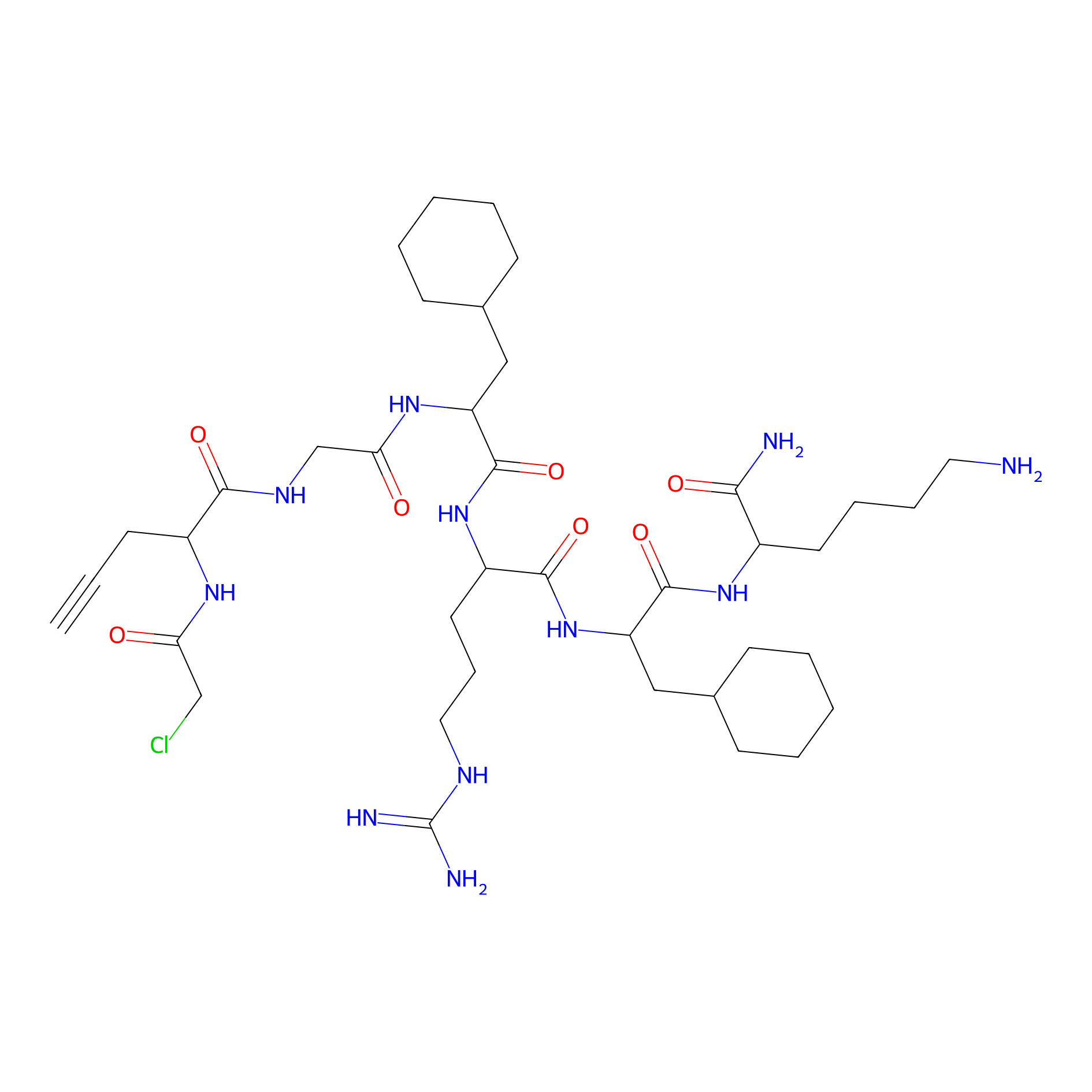 |
N.A. | LDD0428 | [9] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [10] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | H186(0.00); H91(0.00) | LDD0222 | [8] |
| LDCM0116 | HHS-0101 | DM93 | Y92(0.97) | LDD0264 | [6] |
| LDCM0117 | HHS-0201 | DM93 | Y92(0.90) | LDD0265 | [6] |
| LDCM0118 | HHS-0301 | DM93 | Y92(0.89) | LDD0266 | [6] |
| LDCM0119 | HHS-0401 | DM93 | Y92(1.06) | LDD0267 | [6] |
| LDCM0120 | HHS-0701 | DM93 | Y92(1.18) | LDD0268 | [6] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [8] |
| LDCM0022 | KB02 | BRX211 | C153(1.30) | LDD2268 | [5] |
| LDCM0023 | KB03 | BRX211 | C153(1.96) | LDD2685 | [5] |
| LDCM0024 | KB05 | OPM-2 | C55(1.04); C153(1.20) | LDD3377 | [5] |
| LDCM0109 | NEM | HeLa | H91(0.00); H186(0.00) | LDD0223 | [8] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cytoplasmic dynein 1 light intermediate chain 1 (DYNC1LI1) | Dynein light intermediate chain family | Q9Y6G9 | |||
| Clavesin-2 (CLVS2) | . | Q5SYC1 | |||
References
