Details of the Target
General Information of Target
| Target ID | LDTP02177 | |||||
|---|---|---|---|---|---|---|
| Target Name | Acyl-CoA-binding protein (DBI) | |||||
| Gene Name | DBI | |||||
| Gene ID | 1622 | |||||
| Synonyms |
Acyl-CoA-binding protein; ACBP; Diazepam-binding inhibitor; DBI; Endozepine; EP |
|||||
| 3D Structure | ||||||
| Sequence |
MSQAEFEKAAEEVRHLKTKPSDEEMLFIYGHYKQATVGDINTERPGMLDFTGKAKWDAWN
ELKGTSKEDAMKAYINKVEELKKKYGI |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
ACBP family
|
|||||
| Subcellular location |
Endoplasmic reticulum
|
|||||
| Function |
Binds medium- and long-chain acyl-CoA esters with very high affinity and may function as an intracellular carrier of acyl-CoA esters. It is also able to displace diazepam from the benzodiazepine (BZD) recognition site located on the GABA type A receptor. It is therefore possible that this protein also acts as a neuropeptide to modulate the action of the GABA receptor.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
AZ-9 Probe Info |
 |
10.07 | LDD0393 | [1] | |
|
STPyne Probe Info |
 |
K19(1.61); K55(8.36); K77(10.00) | LDD0277 | [2] | |
|
ONAyne Probe Info |
 |
N.A. | LDD0273 | [2] | |
|
HHS-475 Probe Info |
 |
Y29(0.16) | LDD0264 | [3] | |
|
W1 Probe Info |
 |
K55(4.49); W56(4.49) | LDD0237 | [4] | |
|
AMP probe Probe Info |
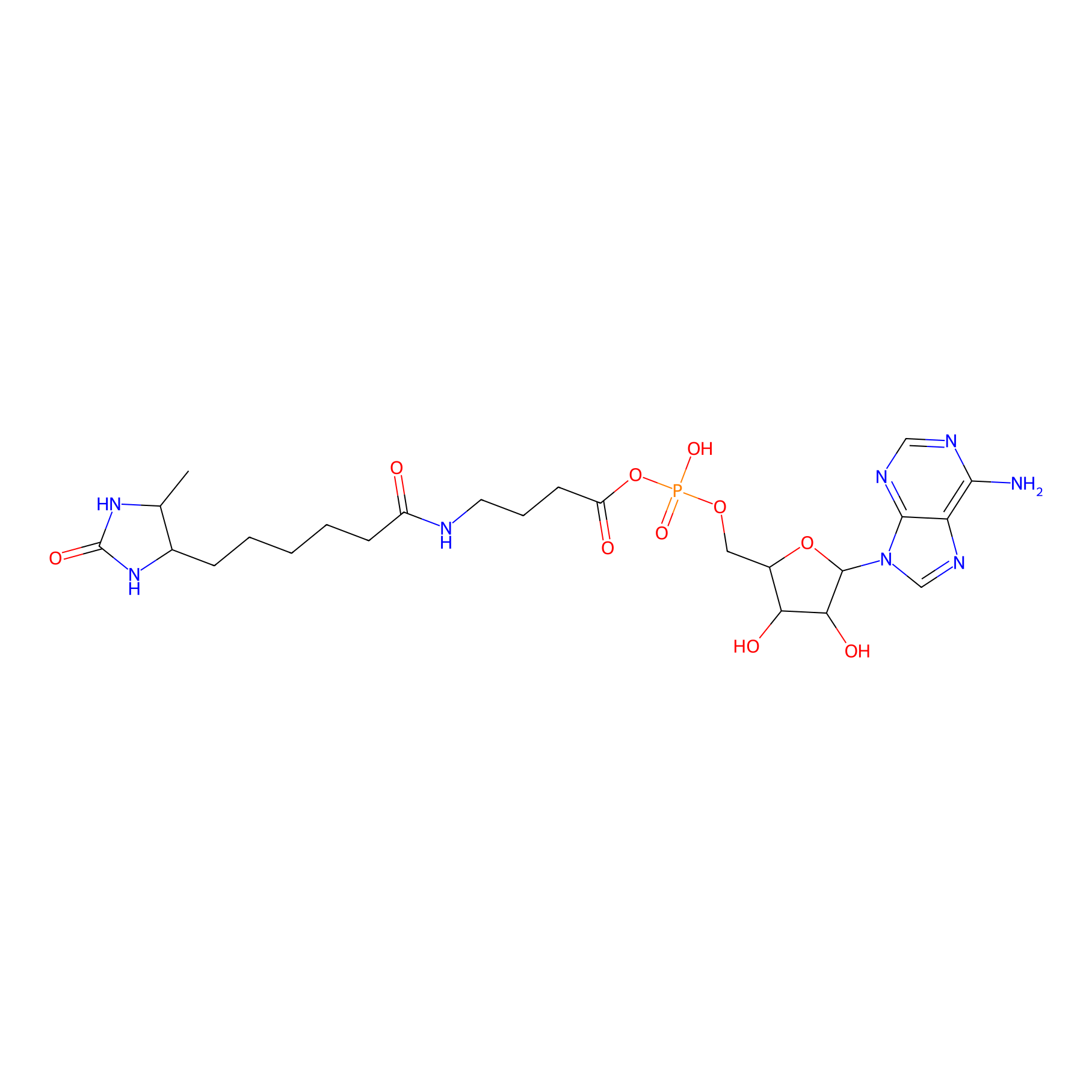 |
N.A. | LDD0200 | [5] | |
|
ATP probe Probe Info |
 |
K55(0.00); K19(0.00); K63(0.00); K67(0.00) | LDD0199 | [5] | |
|
ATP probe Probe Info |
 |
K67(0.00); K82(0.00); K63(0.00) | LDD0035 | [6] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [7] | |
|
SF Probe Info |
 |
Y32(0.00); K33(0.00) | LDD0028 | [8] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0116 | HHS-0101 | DM93 | Y29(0.16) | LDD0264 | [3] |
| LDCM0117 | HHS-0201 | DM93 | Y29(0.77) | LDD0265 | [3] |
| LDCM0118 | HHS-0301 | DM93 | Y29(0.16) | LDD0266 | [3] |
| LDCM0119 | HHS-0401 | DM93 | Y29(1.57) | LDD0267 | [3] |
| LDCM0120 | HHS-0701 | DM93 | Y29(0.07) | LDD0268 | [3] |
| LDCM0110 | W12 | Hep-G2 | K55(4.49); W56(4.49) | LDD0237 | [4] |
| LDCM0111 | W14 | Hep-G2 | K55(3.86) | LDD0238 | [4] |
| LDCM0112 | W16 | Hep-G2 | K55(0.72); W56(0.72) | LDD0239 | [4] |
| LDCM0113 | W17 | Hep-G2 | K55(3.42); W56(3.42) | LDD0240 | [4] |
The Interaction Atlas With This Target
References
