Details of the Target
General Information of Target
| Target ID | LDTP01826 | |||||
|---|---|---|---|---|---|---|
| Target Name | GTPase HRas (HRAS) | |||||
| Gene Name | HRAS | |||||
| Gene ID | 3265 | |||||
| Synonyms |
HRAS1; GTPase HRas; EC 3.6.5.2; H-Ras-1; Ha-Ras; Transforming protein p21; c-H-ras; p21ras) [Cleaved into: GTPase HRas, N-terminally processed] |
|||||
| 3D Structure | ||||||
| Sequence |
MTEYKLVVVGAGGVGKSALTIQLIQNHFVDEYDPTIEDSYRKQVVIDGETCLLDILDTAG
QEEYSAMRDQYMRTGEGFLCVFAINNTKSFEDIHQYREQIKRVKDSDDVPMVLVGNKCDL AARTVESRQAQDLARSYGIPYIETSAKTRQGVEDAFYTLVREIRQHKLRKLNPPDESGPG CMSCKCVLS |
|||||
| Target Type |
Literature-reported
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Small GTPase superfamily, Ras family
|
|||||
| Subcellular location |
Nucleus; Cell membrane
|
|||||
| Function | Involved in the activation of Ras protein signal transduction. Ras proteins bind GDP/GTP and possess intrinsic GTPase activity. | |||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| AN3CA | SNV: p.F82L | . | |||
| HCC1419 | SNV: p.R128W | . | |||
| HS578T | SNV: p.G12D | . | |||
| KASUMI2 | SNV: p.G13S | . | |||
| KYSE30 | SNV: p.Q61L | DBIA Probe Info | |||
| MCC13 | SNV: p.Q61R | . | |||
| MCC26 | SNV: p.G12D | . | |||
| MELJUSO | SNV: p.G13D | . | |||
| MOLT4 | SNV: p.R123H | . | |||
| RL952 | SNV: p.Q61H | . | |||
| T24 | SNV: p.G12V | DBIA Probe Info | |||
| ZR751 | SNV: p.E162K | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
14.07 | LDD0402 | [1] | |
|
Jackson_1 Probe Info |
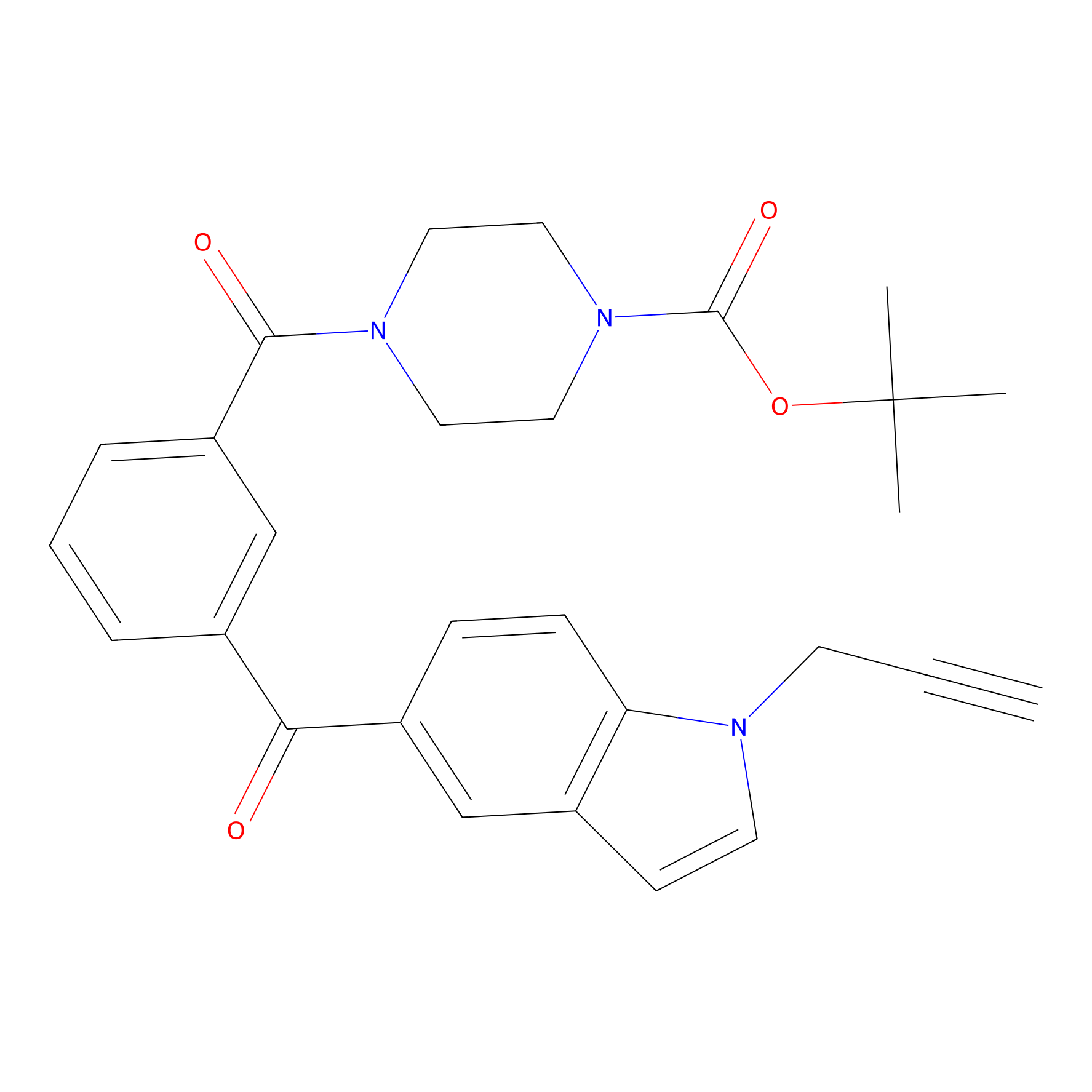 |
20.00 | LDD0122 | [2] | |
|
IPM Probe Info |
 |
C118(1.51) | LDD1701 | [3] | |
|
DBIA Probe Info |
 |
C181(0.97); C184(0.97) | LDD0078 | [4] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C80(0.00); C51(0.00) | LDD0038 | [5] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [5] | |
|
AOyne Probe Info |
 |
14.80 | LDD0443 | [6] | |
|
NAIA_5 Probe Info |
 |
C80(0.00); C118(0.00) | LDD2223 | [7] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0263 | AC143 | HCT 116 | C181(1.52); C184(1.52) | LDD0580 | [4] |
| LDCM0264 | AC144 | HCT 116 | C181(0.57); C184(0.57) | LDD0581 | [4] |
| LDCM0265 | AC145 | HCT 116 | C181(1.48); C184(1.48) | LDD0582 | [4] |
| LDCM0266 | AC146 | HCT 116 | C181(0.88); C184(0.88) | LDD0583 | [4] |
| LDCM0267 | AC147 | HCT 116 | C181(0.94); C184(0.94) | LDD0584 | [4] |
| LDCM0268 | AC148 | HCT 116 | C181(1.58); C184(1.58) | LDD0585 | [4] |
| LDCM0269 | AC149 | HCT 116 | C181(1.01); C184(1.01) | LDD0586 | [4] |
| LDCM0271 | AC150 | HCT 116 | C181(0.85); C184(0.85) | LDD0588 | [4] |
| LDCM0272 | AC151 | HCT 116 | C181(0.86); C184(0.86) | LDD0589 | [4] |
| LDCM0273 | AC152 | HCT 116 | C181(0.95); C184(0.95) | LDD0590 | [4] |
| LDCM0274 | AC153 | HCT 116 | C181(1.25); C184(1.25) | LDD0591 | [4] |
| LDCM0621 | AC154 | HCT 116 | C181(1.31); C184(1.31) | LDD2158 | [4] |
| LDCM0622 | AC155 | HCT 116 | C181(1.20); C184(1.20) | LDD2159 | [4] |
| LDCM0623 | AC156 | HCT 116 | C181(1.06); C184(1.06) | LDD2160 | [4] |
| LDCM0624 | AC157 | HCT 116 | C181(0.93); C184(0.93) | LDD2161 | [4] |
| LDCM0296 | AC35 | HCT 116 | C181(1.09); C184(1.09) | LDD0613 | [4] |
| LDCM0297 | AC36 | HCT 116 | C181(0.86); C184(0.86) | LDD0614 | [4] |
| LDCM0298 | AC37 | HCT 116 | C181(1.02); C184(1.02) | LDD0615 | [4] |
| LDCM0299 | AC38 | HCT 116 | C181(0.91); C184(0.91) | LDD0616 | [4] |
| LDCM0300 | AC39 | HCT 116 | C181(0.99); C184(0.99) | LDD0617 | [4] |
| LDCM0302 | AC40 | HCT 116 | C181(1.00); C184(1.00) | LDD0619 | [4] |
| LDCM0303 | AC41 | HCT 116 | C181(1.10); C184(1.10) | LDD0620 | [4] |
| LDCM0304 | AC42 | HCT 116 | C181(1.05); C184(1.05) | LDD0621 | [4] |
| LDCM0305 | AC43 | HCT 116 | C181(1.10); C184(1.10) | LDD0622 | [4] |
| LDCM0306 | AC44 | HCT 116 | C181(1.23); C184(1.23) | LDD0623 | [4] |
| LDCM0307 | AC45 | HCT 116 | C181(1.22); C184(1.22) | LDD0624 | [4] |
| LDCM0349 | AC83 | HCT 116 | C181(1.03); C184(1.03) | LDD0666 | [4] |
| LDCM0350 | AC84 | HCT 116 | C181(0.84); C184(0.84) | LDD0667 | [4] |
| LDCM0351 | AC85 | HCT 116 | C181(0.91); C184(0.91) | LDD0668 | [4] |
| LDCM0352 | AC86 | HCT 116 | C181(1.12); C184(1.12) | LDD0669 | [4] |
| LDCM0353 | AC87 | HCT 116 | C181(1.16); C184(1.16) | LDD0670 | [4] |
| LDCM0354 | AC88 | HCT 116 | C181(1.25); C184(1.25) | LDD0671 | [4] |
| LDCM0355 | AC89 | HCT 116 | C181(1.26); C184(1.26) | LDD0672 | [4] |
| LDCM0357 | AC90 | HCT 116 | C181(1.42); C184(1.42) | LDD0674 | [4] |
| LDCM0358 | AC91 | HCT 116 | C181(0.95); C184(0.95) | LDD0675 | [4] |
| LDCM0359 | AC92 | HCT 116 | C181(1.10); C184(1.10) | LDD0676 | [4] |
| LDCM0360 | AC93 | HCT 116 | C181(1.07); C184(1.07) | LDD0677 | [4] |
| LDCM0361 | AC94 | HCT 116 | C181(0.99); C184(0.99) | LDD0678 | [4] |
| LDCM0362 | AC95 | HCT 116 | C181(1.72); C184(1.72) | LDD0679 | [4] |
| LDCM0363 | AC96 | HCT 116 | C181(0.87); C184(0.87) | LDD0680 | [4] |
| LDCM0364 | AC97 | HCT 116 | C181(1.24); C184(1.24) | LDD0681 | [4] |
| LDCM0020 | ARS-1620 | HCC44 | C181(0.97); C184(0.97) | LDD0078 | [4] |
| LDCM0372 | CL103 | HEK-293T | C181(1.06) | LDD1576 | [8] |
| LDCM0376 | CL107 | HEK-293T | C181(1.13) | LDD1580 | [8] |
| LDCM0381 | CL111 | HEK-293T | C181(0.95) | LDD1585 | [8] |
| LDCM0385 | CL115 | HEK-293T | C181(1.10) | LDD1589 | [8] |
| LDCM0387 | CL117 | HCT 116 | C181(0.86); C184(0.86) | LDD0704 | [4] |
| LDCM0388 | CL118 | HCT 116 | C181(0.83); C184(0.83) | LDD0705 | [4] |
| LDCM0389 | CL119 | HCT 116 | C181(0.99); C184(0.99) | LDD0706 | [4] |
| LDCM0391 | CL120 | HCT 116 | C181(1.12); C184(1.12) | LDD0708 | [4] |
| LDCM0394 | CL123 | HEK-293T | C181(1.99) | LDD1598 | [8] |
| LDCM0398 | CL127 | HEK-293T | C181(1.43) | LDD1602 | [8] |
| LDCM0402 | CL15 | HEK-293T | C181(1.04) | LDD1606 | [8] |
| LDCM0403 | CL16 | HCT 116 | C181(1.04); C184(1.04) | LDD0720 | [4] |
| LDCM0404 | CL17 | HCT 116 | C181(0.58); C184(0.58) | LDD0721 | [4] |
| LDCM0405 | CL18 | HCT 116 | C181(0.81); C184(0.81) | LDD0722 | [4] |
| LDCM0406 | CL19 | HCT 116 | C181(1.02); C184(1.02) | LDD0723 | [4] |
| LDCM0408 | CL20 | HCT 116 | C181(1.14); C184(1.14) | LDD0725 | [4] |
| LDCM0409 | CL21 | HCT 116 | C181(0.92); C184(0.92) | LDD0726 | [4] |
| LDCM0410 | CL22 | HCT 116 | C181(1.00); C184(1.00) | LDD0727 | [4] |
| LDCM0411 | CL23 | HCT 116 | C181(1.30); C184(1.30) | LDD0728 | [4] |
| LDCM0412 | CL24 | HCT 116 | C181(0.88); C184(0.88) | LDD0729 | [4] |
| LDCM0413 | CL25 | HCT 116 | C181(1.04); C184(1.04) | LDD0730 | [4] |
| LDCM0414 | CL26 | HCT 116 | C181(1.20); C184(1.20) | LDD0731 | [4] |
| LDCM0415 | CL27 | HCT 116 | C181(1.02); C184(1.02) | LDD0732 | [4] |
| LDCM0416 | CL28 | HCT 116 | C181(0.94); C184(0.94) | LDD0733 | [4] |
| LDCM0417 | CL29 | HCT 116 | C181(1.41); C184(1.41) | LDD0734 | [4] |
| LDCM0418 | CL3 | HEK-293T | C181(1.03) | LDD1622 | [8] |
| LDCM0419 | CL30 | HCT 116 | C181(1.17); C184(1.17) | LDD0736 | [4] |
| LDCM0428 | CL39 | HEK-293T | C181(1.05) | LDD1632 | [8] |
| LDCM0453 | CL61 | HCT 116 | C181(0.74); C184(0.74) | LDD0770 | [4] |
| LDCM0454 | CL62 | HCT 116 | C181(1.12); C184(1.12) | LDD0771 | [4] |
| LDCM0455 | CL63 | HCT 116 | C181(0.75); C184(0.75) | LDD0772 | [4] |
| LDCM0456 | CL64 | HCT 116 | C181(0.76); C184(0.76) | LDD0773 | [4] |
| LDCM0457 | CL65 | HCT 116 | C181(0.78); C184(0.78) | LDD0774 | [4] |
| LDCM0458 | CL66 | HCT 116 | C181(0.88); C184(0.88) | LDD0775 | [4] |
| LDCM0459 | CL67 | HCT 116 | C181(0.70); C184(0.70) | LDD0776 | [4] |
| LDCM0460 | CL68 | HCT 116 | C181(0.95); C184(0.95) | LDD0777 | [4] |
| LDCM0461 | CL69 | HCT 116 | C181(0.74); C184(0.74) | LDD0778 | [4] |
| LDCM0463 | CL70 | HCT 116 | C181(0.72); C184(0.72) | LDD0780 | [4] |
| LDCM0464 | CL71 | HCT 116 | C181(0.90); C184(0.90) | LDD0781 | [4] |
| LDCM0465 | CL72 | HCT 116 | C181(0.68); C184(0.68) | LDD0782 | [4] |
| LDCM0466 | CL73 | HCT 116 | C181(0.75); C184(0.75) | LDD0783 | [4] |
| LDCM0467 | CL74 | HCT 116 | C181(0.86); C184(0.86) | LDD0784 | [4] |
| LDCM0469 | CL76 | HCT 116 | C181(1.02); C184(1.02) | LDD0786 | [4] |
| LDCM0470 | CL77 | HCT 116 | C181(0.84); C184(0.84) | LDD0787 | [4] |
| LDCM0471 | CL78 | HCT 116 | C181(1.06); C184(1.06) | LDD0788 | [4] |
| LDCM0472 | CL79 | HCT 116 | C181(1.04); C184(1.04) | LDD0789 | [4] |
| LDCM0474 | CL80 | HCT 116 | C181(0.91); C184(0.91) | LDD0791 | [4] |
| LDCM0475 | CL81 | HCT 116 | C181(0.87); C184(0.87) | LDD0792 | [4] |
| LDCM0476 | CL82 | HCT 116 | C181(0.98); C184(0.98) | LDD0793 | [4] |
| LDCM0477 | CL83 | HCT 116 | C181(0.96); C184(0.96) | LDD0794 | [4] |
| LDCM0478 | CL84 | HCT 116 | C181(0.88); C184(0.88) | LDD0795 | [4] |
| LDCM0479 | CL85 | HCT 116 | C181(1.01); C184(1.01) | LDD0796 | [4] |
| LDCM0480 | CL86 | HCT 116 | C181(0.98); C184(0.98) | LDD0797 | [4] |
| LDCM0481 | CL87 | HCT 116 | C181(0.84); C184(0.84) | LDD0798 | [4] |
| LDCM0482 | CL88 | HCT 116 | C181(0.89); C184(0.89) | LDD0799 | [4] |
| LDCM0483 | CL89 | HCT 116 | C181(1.04); C184(1.04) | LDD0800 | [4] |
| LDCM0485 | CL90 | HCT 116 | C181(0.79); C184(0.79) | LDD0802 | [4] |
| LDCM0494 | CL99 | HEK-293T | C181(1.23) | LDD1697 | [8] |
| LDCM0495 | E2913 | HEK-293T | C181(1.05) | LDD1698 | [8] |
| LDCM0468 | Fragment33 | HCT 116 | C181(0.73); C184(0.73) | LDD0785 | [4] |
| LDCM0022 | KB02 | HEK-293T | C51(0.89); C181(0.85) | LDD1492 | [8] |
| LDCM0023 | KB03 | HEK-293T | C51(0.90); C181(1.05) | LDD1497 | [8] |
| LDCM0024 | KB05 | MIA PaCa-2 | C118(1.08) | LDD3330 | [9] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
Other
The Drug(s) Related To This Target
Investigative
Discontinued
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Isis 2503 | Antisense drug | D01XDN | |||
References
