Details of the Target
General Information of Target
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C13(2.93) | LDD3419 | [1] | |
|
Alkyne-RA190 Probe Info |
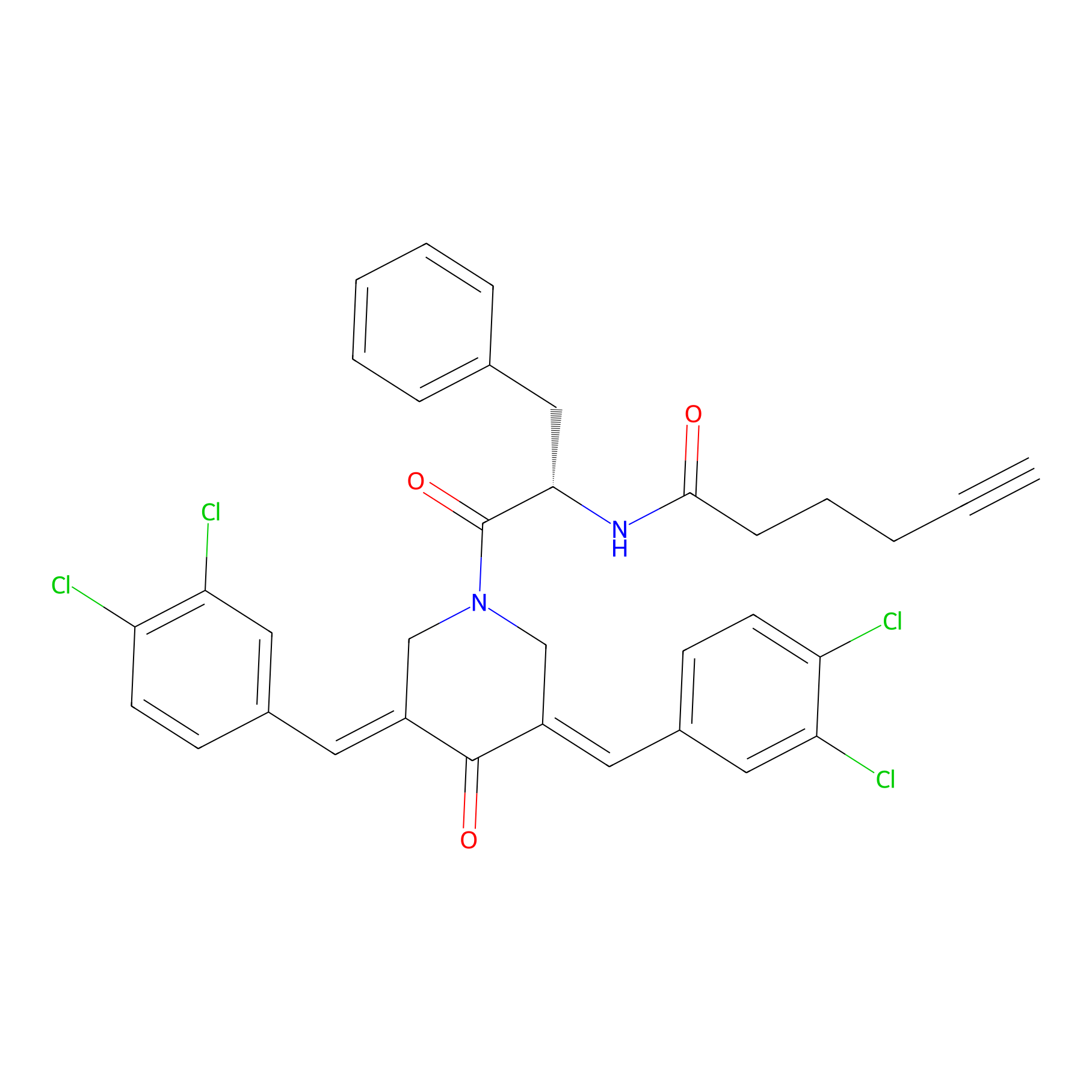 |
2.10 | LDD0303 | [2] | |
|
IA-alkyne Probe Info |
 |
C219(5.09) | LDD1706 | [3] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0216 | AC100 | PaTu 8988t | C143(1.12); C149(1.12) | LDD1095 | [4] |
| LDCM0217 | AC101 | PaTu 8988t | C143(1.15); C149(1.15) | LDD1096 | [4] |
| LDCM0218 | AC102 | PaTu 8988t | C143(1.11); C149(1.11) | LDD1097 | [4] |
| LDCM0219 | AC103 | PaTu 8988t | C143(1.21); C149(1.21) | LDD1098 | [4] |
| LDCM0220 | AC104 | PaTu 8988t | C143(1.06); C149(1.06) | LDD1099 | [4] |
| LDCM0221 | AC105 | PaTu 8988t | C143(1.12); C149(1.12) | LDD1100 | [4] |
| LDCM0222 | AC106 | PaTu 8988t | C143(1.28); C149(1.28) | LDD1101 | [4] |
| LDCM0223 | AC107 | PaTu 8988t | C143(1.05); C149(1.05) | LDD1102 | [4] |
| LDCM0224 | AC108 | PaTu 8988t | C143(1.08); C149(1.08) | LDD1103 | [4] |
| LDCM0225 | AC109 | PaTu 8988t | C143(1.29); C149(1.29) | LDD1104 | [4] |
| LDCM0227 | AC110 | PaTu 8988t | C143(1.11); C149(1.11) | LDD1106 | [4] |
| LDCM0228 | AC111 | PaTu 8988t | C143(1.50); C149(1.50) | LDD1107 | [4] |
| LDCM0229 | AC112 | PaTu 8988t | C143(1.12); C149(1.12) | LDD1108 | [4] |
| LDCM0230 | AC113 | PaTu 8988t | C143(0.93); C149(0.93) | LDD1109 | [4] |
| LDCM0231 | AC114 | PaTu 8988t | C143(1.03); C149(1.03) | LDD1110 | [4] |
| LDCM0232 | AC115 | PaTu 8988t | C143(1.24); C149(1.24) | LDD1111 | [4] |
| LDCM0233 | AC116 | PaTu 8988t | C143(0.91); C149(0.91) | LDD1112 | [4] |
| LDCM0234 | AC117 | PaTu 8988t | C143(0.88); C149(0.88) | LDD1113 | [4] |
| LDCM0235 | AC118 | PaTu 8988t | C143(1.39); C149(1.39) | LDD1114 | [4] |
| LDCM0236 | AC119 | PaTu 8988t | C143(0.89); C149(0.89) | LDD1115 | [4] |
| LDCM0238 | AC120 | PaTu 8988t | C143(0.70); C149(0.70) | LDD1117 | [4] |
| LDCM0239 | AC121 | PaTu 8988t | C143(0.80); C149(0.80) | LDD1118 | [4] |
| LDCM0240 | AC122 | PaTu 8988t | C143(0.89); C149(0.89) | LDD1119 | [4] |
| LDCM0241 | AC123 | PaTu 8988t | C143(0.73); C149(0.73) | LDD1120 | [4] |
| LDCM0242 | AC124 | PaTu 8988t | C143(1.05); C149(1.05) | LDD1121 | [4] |
| LDCM0243 | AC125 | PaTu 8988t | C143(1.53); C149(1.53) | LDD1122 | [4] |
| LDCM0244 | AC126 | PaTu 8988t | C143(1.07); C149(1.07) | LDD1123 | [4] |
| LDCM0245 | AC127 | PaTu 8988t | C143(0.91); C149(0.91) | LDD1124 | [4] |
| LDCM0246 | AC128 | PaTu 8988t | C143(1.17); C149(1.17) | LDD1125 | [4] |
| LDCM0247 | AC129 | PaTu 8988t | C143(1.09); C149(1.09) | LDD1126 | [4] |
| LDCM0249 | AC130 | PaTu 8988t | C143(0.95); C149(0.95) | LDD1128 | [4] |
| LDCM0250 | AC131 | PaTu 8988t | C143(1.15); C149(1.15) | LDD1129 | [4] |
| LDCM0251 | AC132 | PaTu 8988t | C143(1.00); C149(1.00) | LDD1130 | [4] |
| LDCM0252 | AC133 | PaTu 8988t | C143(1.18); C149(1.18) | LDD1131 | [4] |
| LDCM0253 | AC134 | PaTu 8988t | C143(0.97); C149(0.97) | LDD1132 | [4] |
| LDCM0254 | AC135 | PaTu 8988t | C143(1.04); C149(1.04) | LDD1133 | [4] |
| LDCM0255 | AC136 | PaTu 8988t | C143(1.15); C149(1.15) | LDD1134 | [4] |
| LDCM0256 | AC137 | PaTu 8988t | C143(1.13); C149(1.13) | LDD1135 | [4] |
| LDCM0257 | AC138 | PaTu 8988t | C143(1.09); C149(1.09) | LDD1136 | [4] |
| LDCM0258 | AC139 | PaTu 8988t | C143(1.07); C149(1.07) | LDD1137 | [4] |
| LDCM0260 | AC140 | PaTu 8988t | C143(1.19); C149(1.19) | LDD1139 | [4] |
| LDCM0261 | AC141 | PaTu 8988t | C143(1.12); C149(1.12) | LDD1140 | [4] |
| LDCM0262 | AC142 | PaTu 8988t | C143(1.15); C149(1.15) | LDD1141 | [4] |
| LDCM0263 | AC143 | PaTu 8988t | C143(1.06); C149(1.06) | LDD1142 | [4] |
| LDCM0264 | AC144 | PaTu 8988t | C143(1.12); C149(1.12) | LDD1143 | [4] |
| LDCM0265 | AC145 | PaTu 8988t | C143(1.24); C149(1.24) | LDD1144 | [4] |
| LDCM0266 | AC146 | PaTu 8988t | C143(1.00); C149(1.00) | LDD1145 | [4] |
| LDCM0267 | AC147 | PaTu 8988t | C143(1.26); C149(1.26) | LDD1146 | [4] |
| LDCM0268 | AC148 | PaTu 8988t | C143(1.00); C149(1.00) | LDD1147 | [4] |
| LDCM0269 | AC149 | PaTu 8988t | C143(1.28); C149(1.28) | LDD1148 | [4] |
| LDCM0271 | AC150 | PaTu 8988t | C143(1.57); C149(1.57) | LDD1150 | [4] |
| LDCM0272 | AC151 | PaTu 8988t | C143(1.77); C149(1.77) | LDD1151 | [4] |
| LDCM0273 | AC152 | PaTu 8988t | C143(1.52); C149(1.52) | LDD1152 | [4] |
| LDCM0274 | AC153 | PaTu 8988t | C143(1.75); C149(1.75) | LDD1153 | [4] |
| LDCM0621 | AC154 | PaTu 8988t | C143(2.17); C149(2.17) | LDD2166 | [4] |
| LDCM0622 | AC155 | PaTu 8988t | C143(1.87); C149(1.87) | LDD2167 | [4] |
| LDCM0623 | AC156 | PaTu 8988t | C143(1.62); C149(1.62) | LDD2168 | [4] |
| LDCM0624 | AC157 | PaTu 8988t | C143(1.71); C149(1.71) | LDD2169 | [4] |
| LDCM0276 | AC17 | PaTu 8988t | C143(1.39); C149(1.39) | LDD1155 | [4] |
| LDCM0277 | AC18 | PaTu 8988t | C143(0.87); C149(0.87) | LDD1156 | [4] |
| LDCM0278 | AC19 | PaTu 8988t | C143(0.93); C149(0.93) | LDD1157 | [4] |
| LDCM0280 | AC20 | PaTu 8988t | C143(0.98); C149(0.98) | LDD1159 | [4] |
| LDCM0281 | AC21 | PaTu 8988t | C143(0.95); C149(0.95) | LDD1160 | [4] |
| LDCM0282 | AC22 | PaTu 8988t | C143(1.10); C149(1.10) | LDD1161 | [4] |
| LDCM0283 | AC23 | PaTu 8988t | C143(1.03); C149(1.03) | LDD1162 | [4] |
| LDCM0284 | AC24 | PaTu 8988t | C143(1.08); C149(1.08) | LDD1163 | [4] |
| LDCM0365 | AC98 | PaTu 8988t | C143(1.20); C149(1.20) | LDD1244 | [4] |
| LDCM0366 | AC99 | PaTu 8988t | C143(1.09); C149(1.09) | LDD1245 | [4] |
| LDCM0374 | CL105 | PaTu 8988t | C143(0.83); C149(0.83) | LDD1253 | [4] |
| LDCM0375 | CL106 | PaTu 8988t | C143(1.15); C149(1.15) | LDD1254 | [4] |
| LDCM0376 | CL107 | PaTu 8988t | C143(1.05); C149(1.05) | LDD1255 | [4] |
| LDCM0377 | CL108 | PaTu 8988t | C143(0.83); C149(0.83) | LDD1256 | [4] |
| LDCM0378 | CL109 | PaTu 8988t | C143(1.14); C149(1.14) | LDD1257 | [4] |
| LDCM0380 | CL110 | PaTu 8988t | C143(1.26); C149(1.26) | LDD1259 | [4] |
| LDCM0381 | CL111 | PaTu 8988t | C143(1.25); C149(1.25) | LDD1260 | [4] |
| LDCM0420 | CL31 | PaTu 8988t | C143(1.19); C149(1.19) | LDD1299 | [4] |
| LDCM0421 | CL32 | PaTu 8988t | C143(1.76); C149(1.76) | LDD1300 | [4] |
| LDCM0422 | CL33 | PaTu 8988t | C143(1.67); C149(1.67) | LDD1301 | [4] |
| LDCM0423 | CL34 | PaTu 8988t | C143(1.22); C149(1.22) | LDD1302 | [4] |
| LDCM0424 | CL35 | PaTu 8988t | C143(1.43); C149(1.43) | LDD1303 | [4] |
| LDCM0425 | CL36 | PaTu 8988t | C143(1.23); C149(1.23) | LDD1304 | [4] |
| LDCM0426 | CL37 | PaTu 8988t | C143(1.86); C149(1.86) | LDD1305 | [4] |
| LDCM0428 | CL39 | PaTu 8988t | C143(1.79); C149(1.79) | LDD1307 | [4] |
| LDCM0430 | CL40 | PaTu 8988t | C143(1.63); C149(1.63) | LDD1309 | [4] |
| LDCM0431 | CL41 | PaTu 8988t | C143(1.49); C149(1.49) | LDD1310 | [4] |
| LDCM0432 | CL42 | PaTu 8988t | C143(1.93); C149(1.93) | LDD1311 | [4] |
| LDCM0433 | CL43 | PaTu 8988t | C143(2.27); C149(2.27) | LDD1312 | [4] |
| LDCM0434 | CL44 | PaTu 8988t | C143(1.53); C149(1.53) | LDD1313 | [4] |
| LDCM0435 | CL45 | PaTu 8988t | C143(2.04); C149(2.04) | LDD1314 | [4] |
| LDCM0469 | CL76 | PaTu 8988t | C143(1.82); C149(1.82) | LDD1348 | [4] |
| LDCM0470 | CL77 | PaTu 8988t | C143(1.52); C149(1.52) | LDD1349 | [4] |
| LDCM0471 | CL78 | PaTu 8988t | C143(1.01); C149(1.01) | LDD1350 | [4] |
| LDCM0472 | CL79 | PaTu 8988t | C143(0.96); C149(0.96) | LDD1351 | [4] |
| LDCM0474 | CL80 | PaTu 8988t | C143(1.19); C149(1.19) | LDD1353 | [4] |
| LDCM0475 | CL81 | PaTu 8988t | C143(1.02); C149(1.02) | LDD1354 | [4] |
| LDCM0476 | CL82 | PaTu 8988t | C143(0.98); C149(0.98) | LDD1355 | [4] |
| LDCM0477 | CL83 | PaTu 8988t | C143(1.12); C149(1.12) | LDD1356 | [4] |
| LDCM0478 | CL84 | PaTu 8988t | C143(1.15); C149(1.15) | LDD1357 | [4] |
| LDCM0479 | CL85 | PaTu 8988t | C143(1.06); C149(1.06) | LDD1358 | [4] |
| LDCM0480 | CL86 | PaTu 8988t | C143(2.10); C149(2.10) | LDD1359 | [4] |
| LDCM0481 | CL87 | PaTu 8988t | C143(1.34); C149(1.34) | LDD1360 | [4] |
| LDCM0482 | CL88 | PaTu 8988t | C143(1.29); C149(1.29) | LDD1361 | [4] |
| LDCM0483 | CL89 | PaTu 8988t | C143(0.96); C149(0.96) | LDD1362 | [4] |
| LDCM0485 | CL90 | PaTu 8988t | C143(1.14); C149(1.14) | LDD1364 | [4] |
| LDCM0427 | Fragment51 | PaTu 8988t | C143(1.72); C149(1.72) | LDD1306 | [4] |
| LDCM0022 | KB02 | BRX211 | C13(2.55); C219(1.74) | LDD2268 | [1] |
| LDCM0023 | KB03 | BRX211 | C13(5.87) | LDD2685 | [1] |
| LDCM0024 | KB05 | SH-4 | C13(2.93) | LDD3419 | [1] |
| LDCM0131 | RA190 | SK-MEL-5 | 2.10 | LDD0303 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cytochrome c oxidase subunit 5B, mitochondrial (COX5B) | Cytochrome c oxidase subunit 5B family | P10606 | |||
| Cellular tumor antigen p53 (TP53) | P53 family | P04637 | |||
Transcription factor
Other
References
