Details of the Target
General Information of Target
| Target ID | LDTP18547 | |||||
|---|---|---|---|---|---|---|
| Target Name | Protein CDV3 homolog (CDV3) | |||||
| Gene Name | CDV3 | |||||
| Gene ID | 55573 | |||||
| Synonyms |
H41; Protein CDV3 homolog |
|||||
| 3D Structure | ||||||
| Sequence |
MAEIHTPYSSLKKLLSLLNGFVAVSGIILVGLGIGGKCGGASLTNVLGLSSAYLLHVGNL
CLVMGCITVLLGCAGWYGATKESRGTLLFCILSMVIVLIMEVTAATVVLLFFPIVGDVAL EHTFVTLRKNYRGYNEPDDYSTQWNLVMEKLKCCGVNNYTDFSGSSFEMTTGHTYPRSCC KSIGSVSCDGRDVSPNVIHQKGCFHKLLKITKTQSFTLSGSSLGAAVIQRWGSRYVAQAG LELLA |
|||||
| Target Bioclass |
Other
|
|||||
| Family |
CDV3 family
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HT115 | SNV: p.E212D | . | |||
| NCIH1993 | SNV: p.E6G; p.N11H | ATP probe Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
AZ-9 Probe Info |
 |
10.00 | LDD0393 | [1] | |
|
STPyne Probe Info |
 |
K15(10.00); K181(6.20); K203(4.60); K232(7.69) | LDD0277 | [2] | |
|
OPA-S-S-alkyne Probe Info |
 |
K15(2.40) | LDD3494 | [3] | |
|
Probe 1 Probe Info |
 |
Y95(14.30); Y190(82.20) | LDD3495 | [4] | |
|
HHS-475 Probe Info |
 |
Y244(0.87); Y190(0.95); Y169(1.52) | LDD0264 | [5] | |
|
ATP probe Probe Info |
 |
K181(0.00); K15(0.00) | LDD0199 | [6] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [7] | |
|
SF Probe Info |
 |
Y190(0.00); Y95(0.00) | LDD0028 | [8] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [9] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [10] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0357 | [10] | |
|
HHS-465 Probe Info |
 |
Y169(0.00); Y190(0.00); Y244(0.00) | LDD2240 | [11] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
C361 Probe Info |
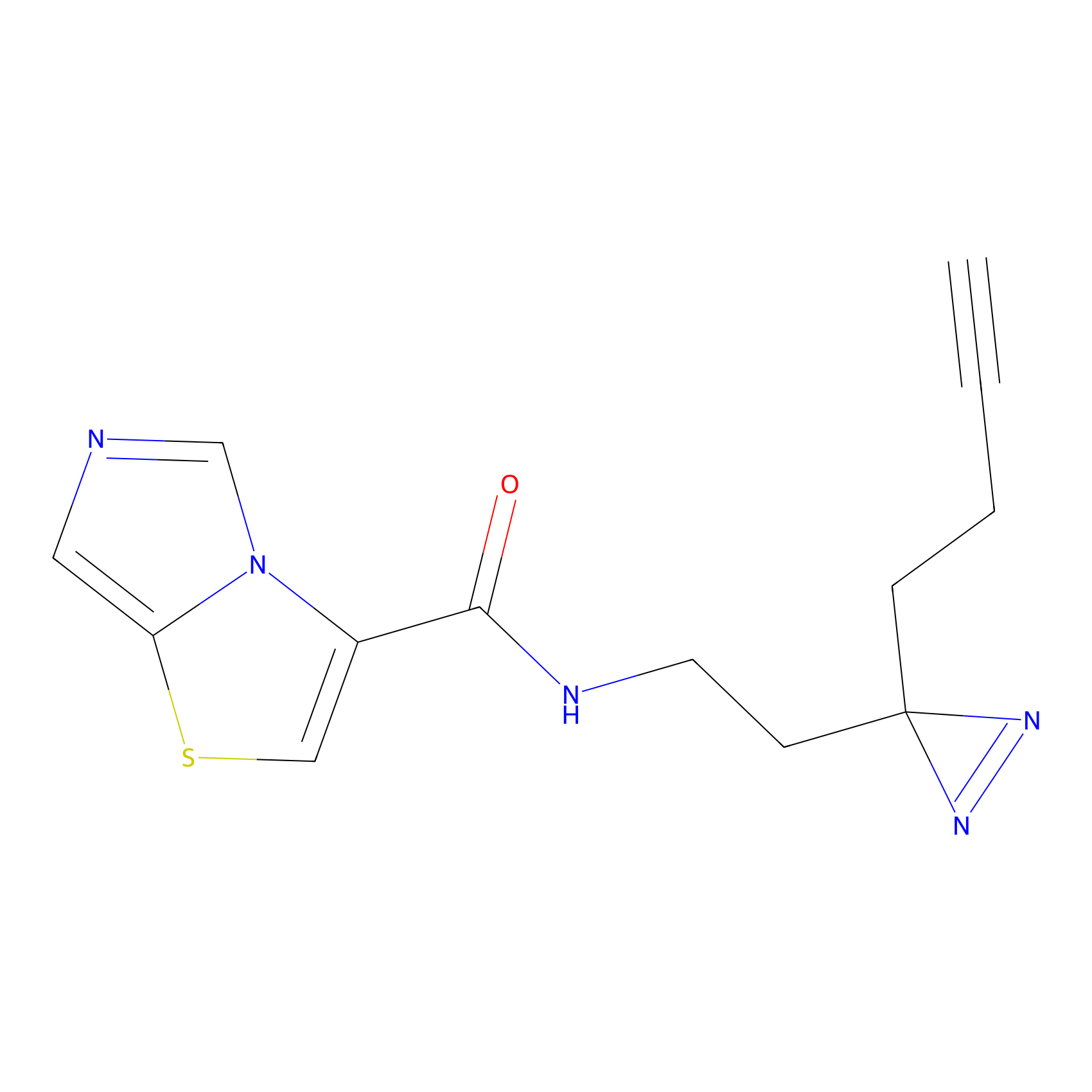 |
17.88 | LDD2022 | [12] | |
|
VE-P Probe Info |
 |
N.A. | LDD0396 | [13] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0116 | HHS-0101 | DM93 | Y244(0.87); Y190(0.95); Y169(1.52) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y244(0.98); Y169(1.11); Y190(1.18) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y169(0.85); Y244(0.97); Y190(1.80) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y244(0.92); Y169(1.00); Y190(1.46) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y190(0.46); Y244(0.79); Y169(1.11) | LDD0268 | [5] |
References
