Details of the Target
General Information of Target
| Target ID | LDTP13752 | |||||
|---|---|---|---|---|---|---|
| Target Name | Cyclin-dependent kinase 11A (CDK11A) | |||||
| Gene Name | CDK11A | |||||
| Gene ID | 728642 | |||||
| Synonyms |
CDC2L2; CDC2L3; PITSLREB; Cyclin-dependent kinase 11A; EC 2.7.11.22; Cell division cycle 2-like protein kinase 2; Cell division protein kinase 11A; Galactosyltransferase-associated protein kinase p58/GTA; PITSLRE serine/threonine-protein kinase CDC2L2
|
|||||
| 3D Structure | ||||||
| Sequence |
MGIQGLLQFIKEASEPIHVRKYKGQVVAVDTYCWLHKGAIACAEKLAKGEPTDRYVGFCM
KFVNMLLSHGIKPILVFDGCTLPSKKEVERSRRERRQANLLKGKQLLREGKVSEARECFT RSINITHAMAHKVIKAARSQGVDCLVAPYEADAQLAYLNKAGIVQAIITEDSDLLAFGCK KVILKMDQFGNGLEIDQARLGMCRQLGDVFTEEKFRYMCILSGCDYLSSLRGIGLAKACK VLRLANNPDIVKVIKKIGHYLKMNITVPEDYINGFIRANNTFLYQLVFDPIKRKLIPLNA YEDDVDPETLSYAGQYVDDSIALQIALGNKDINTFEQIDDYNPDTAMPAHSRSHSWDDKT CQKSANVSSIWHRNYSPRPESGTVSDAPQLKENPSTVGVERVISTKGLNLPRKSSIVKRP RSAELSEDDLLSQYSLSFTKKTKKNSSEGNKSLSFSEVFVPDLVNGPTNKKSVSTPPRTR NKFATFLQRKNEESGAVVVPGTRSRFFCSSDSTDCVSNKVSIQPLDETAVTDKENNLHES EYGDQEGKRLVDTDVARNSSDDIPNNHIPGDHIPDKATVFTDEESYSFESSKFTRTISPP TLGTLRSCFSWSGGLGDFSRTPSPSPSTALQQFRRKSDSPTSLPENNMSDVSQLKSEESS DDESHPLREEACSSQSQESGEFSLQSSNASKLSQCSSKDSDSEESDCNIKLLDSQSDQTS KLRLSHFSKKDTPLRNKVPGLYKSSSADSLSTTKIKPLGPARASGLSKKPASIQKRKHHN AENKPGLQIKLNELWKNFGFKKDSEKLPPCKKPLSPVRDNIQLTPEAEEDIFNKPECGRV QRAIFQ |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein kinase superfamily, CMGC Ser/Thr protein kinase family, CDC2/CDKX subfamily
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Appears to play multiple roles in cell cycle progression, cytokinesis and apoptosis. The p110 isoforms have been suggested to be involved in pre-mRNA splicing, potentially by phosphorylating the splicing protein SFRS7. The p58 isoform may act as a negative regulator of normal cell cycle progression.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K455(1.02); K96(6.67) | LDD0277 | [1] | |
|
Alkylaryl probe 3 Probe Info |
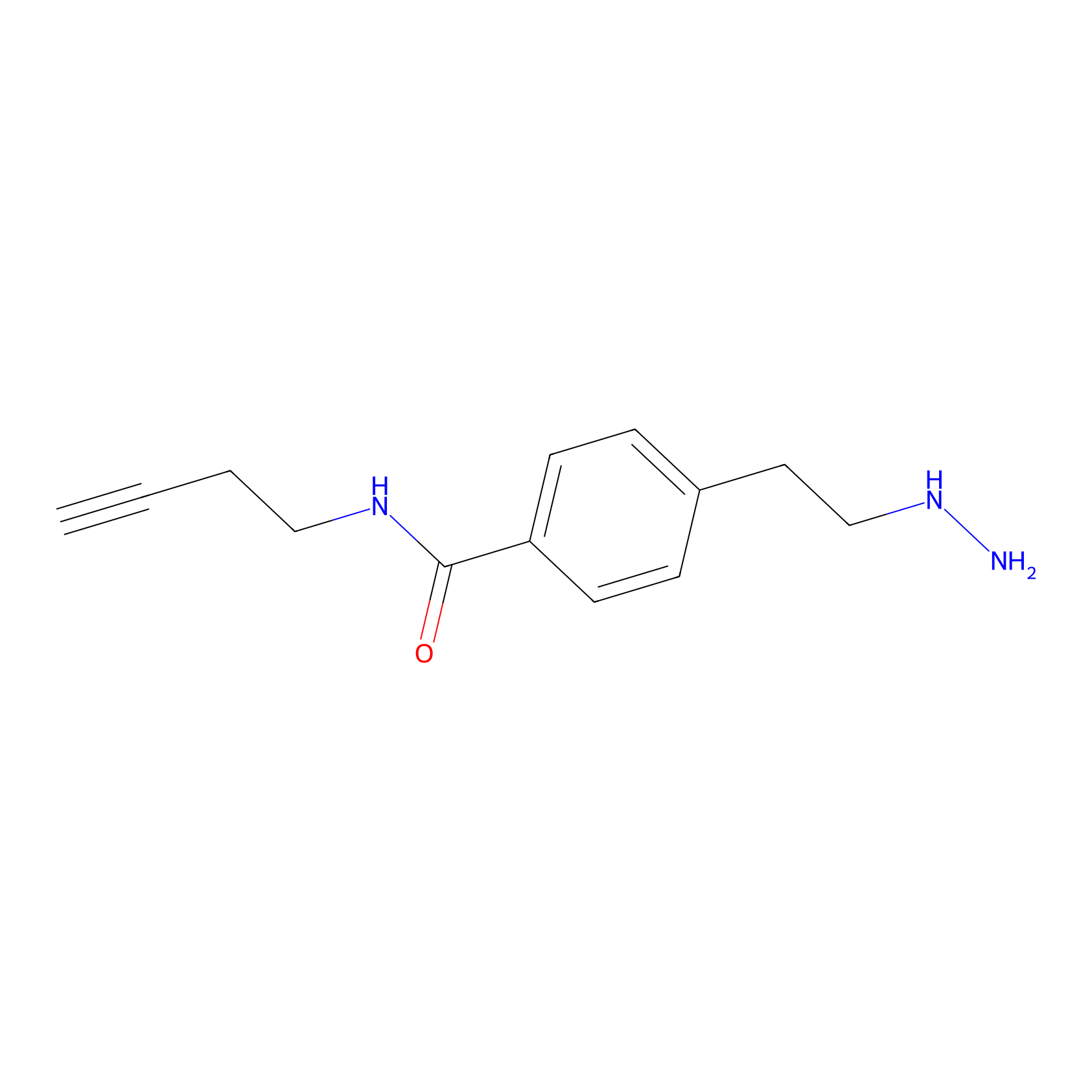 |
12.36 | LDD0382 | [2] | |
|
AHL-Pu-1 Probe Info |
 |
C428(2.01) | LDD0168 | [3] | |
|
HHS-475 Probe Info |
 |
Y441(0.82) | LDD0264 | [4] | |
|
HHS-465 Probe Info |
 |
Y669(10.00) | LDD2237 | [5] | |
|
DBIA Probe Info |
 |
C428(1.13); C420(0.92); C57(0.91) | LDD0078 | [6] | |
|
IA-alkyne Probe Info |
 |
C428(0.00); C420(0.00) | LDD0162 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [8] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [9] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [10] | |
|
NAIA_5 Probe Info |
 |
C57(0.00); C428(0.00); C440(0.00) | LDD2224 | [10] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [11] | |
|
WYneN Probe Info |
 |
N.A. | LDD0021 | [12] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [12] | |
|
ENE Probe Info |
 |
N.A. | LDD0006 | [12] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [12] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [12] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [13] | |
|
VSF Probe Info |
 |
N.A. | LDD0007 | [12] | |
|
Phosphinate-6 Probe Info |
 |
C428(0.00); C420(0.00) | LDD0018 | [14] | |
|
Acrolein Probe Info |
 |
C428(0.00); C57(0.00); H56(0.00); H560(0.00) | LDD0217 | [15] | |
|
Crotonaldehyde Probe Info |
 |
C428(0.00); C420(0.00) | LDD0219 | [15] | |
|
Methacrolein Probe Info |
 |
C428(0.00); C420(0.00) | LDD0218 | [15] | |
|
W1 Probe Info |
 |
C420(0.00); C428(0.00) | LDD0236 | [16] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0025 | 4SU-RNA | HEK-293T | C428(2.01) | LDD0168 | [3] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C428(2.97) | LDD0169 | [3] |
| LDCM0020 | ARS-1620 | HCC44 | C428(1.13); C420(0.92); C57(0.91) | LDD0078 | [6] |
| LDCM0108 | Chloroacetamide | HeLa | C428(0.00); C57(0.00) | LDD0222 | [15] |
| LDCM0632 | CL-Sc | Hep-G2 | C57(1.18) | LDD2227 | [10] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C420(2.18); C428(1.75); C57(1.35) | LDD1702 | [17] |
| LDCM0116 | HHS-0101 | DM93 | Y441(0.82) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y441(0.69) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y441(0.57) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y441(0.59) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y441(0.69) | LDD0268 | [4] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [15] |
| LDCM0023 | KB03 | Jurkat | C420(9.99) | LDD0209 | [18] |
| LDCM0109 | NEM | HeLa | H560(0.00); H482(0.00) | LDD0223 | [15] |
| LDCM0099 | Phenelzine | HEK-293T | 12.36 | LDD0382 | [2] |
The Interaction Atlas With This Target
References
