Details of the Target
General Information of Target
| Target ID | LDTP13533 | |||||
|---|---|---|---|---|---|---|
| Target Name | Long-chain-fatty-acid--CoA ligase 5 (ACSL5) | |||||
| Gene Name | ACSL5 | |||||
| Gene ID | 51703 | |||||
| Synonyms |
ACS5; FACL5; Long-chain-fatty-acid--CoA ligase 5; EC 6.2.1.3; Arachidonate--CoA ligase; EC 6.2.1.15; Long-chain acyl-CoA synthetase 5; LACS 5 |
|||||
| 3D Structure | ||||||
| Sequence |
MFKKFDEKENVSNCIQLKTSVIKGIKNQLIEQFPGIEPWLNQIMPKKDPVKIVRCHEHIE
ILTVNGELLFFRQREGPFYPTLRLLHKYPFILPHQQVDKGAIKFVLSGANIMCPGLTSPG AKLYPAAVDTIVAIMAEGKQHALCVGVMKMSAEDIEKVNKGIGIENIHYLNDGLWHMKTY K |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
ATP-dependent AMP-binding enzyme family
|
|||||
| Subcellular location |
Mitochondrion
|
|||||
| Function |
Catalyzes the conversion of long-chain fatty acids to their active form acyl-CoAs for both synthesis of cellular lipids, and degradation via beta-oxidation. ACSL5 may activate fatty acids from exogenous sources for the synthesis of triacylglycerol destined for intracellular storage. Utilizes a wide range of saturated fatty acids with a preference for C16-C18 unsaturated fatty acids. It was suggested that it may also stimulate fatty acid oxidation. At the villus tip of the crypt-villus axis of the small intestine may sensitize epithelial cells to apoptosis specifically triggered by the death ligand TRAIL. May have a role in the survival of glioma cells.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| HT115 | SNV: p.I273T | DBIA Probe Info | |||
| MCC26 | SNV: p.V439M; p.E559Q | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
YN-1 Probe Info |
 |
100.00 | LDD0444 | [1] | |
|
STPyne Probe Info |
 |
K344(4.93); K361(1.09); K546(0.38); K628(10.00) | LDD0277 | [2] | |
|
ONAyne Probe Info |
 |
K361(3.48) | LDD0275 | [2] | |
|
IPM Probe Info |
 |
C69(0.00); C121(0.00); C70(0.00); C322(0.00) | LDD0241 | [3] | |
|
DBIA Probe Info |
 |
C177(2.54); C93(2.75); C666(1.87); C378(2.64) | LDD3311 | [4] | |
|
BTD Probe Info |
 |
C121(1.15) | LDD2090 | [5] | |
|
Alkyne-RA190 Probe Info |
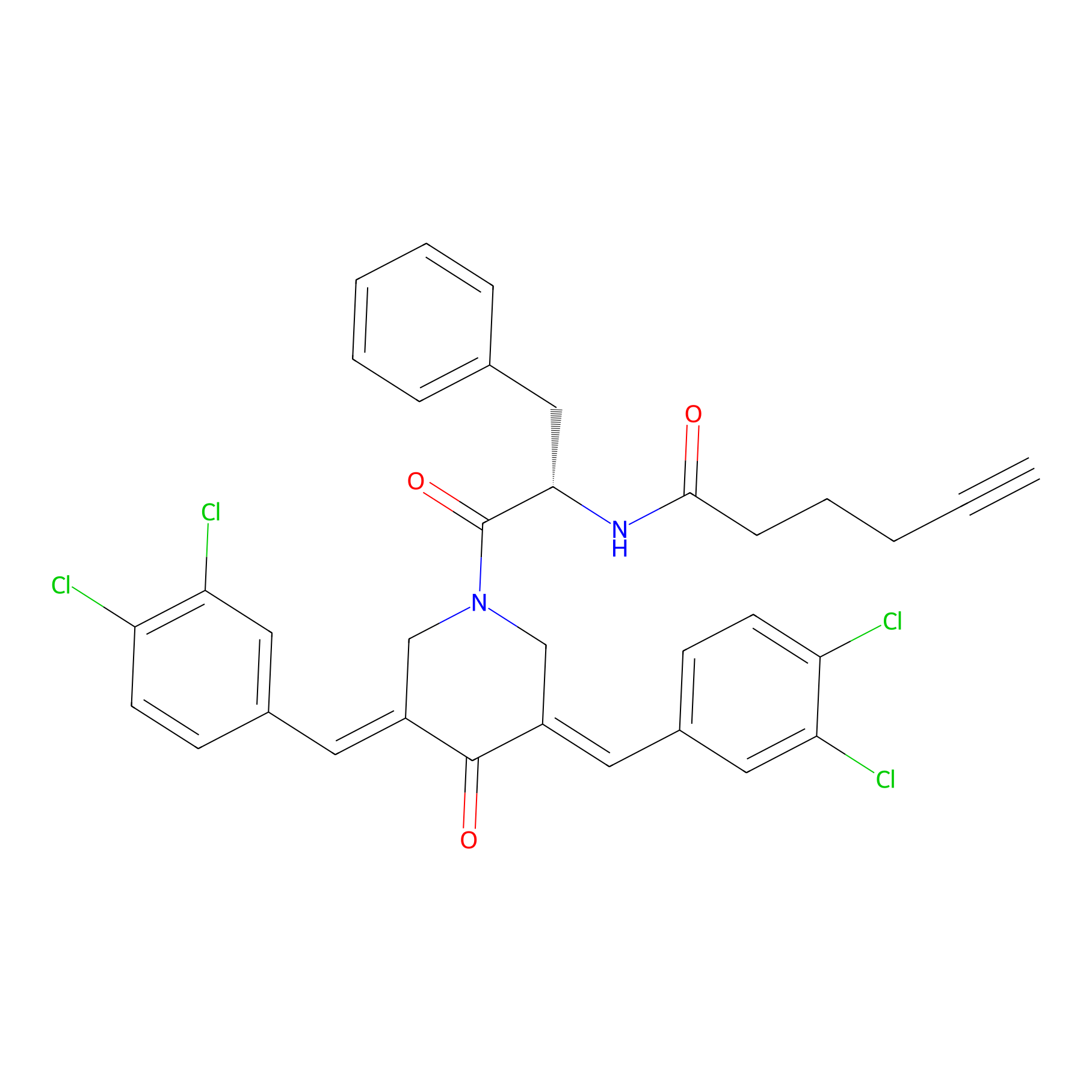 |
2.07 | LDD0299 | [6] | |
|
IA-alkyne Probe Info |
 |
C322(1.02) | LDD0346 | [7] | |
|
IPIAA_H Probe Info |
 |
N.A. | LDD0030 | [8] | |
|
NAIA_4 Probe Info |
 |
C259(0.00); C288(0.00); C322(0.00) | LDD2226 | [9] | |
|
W1 Probe Info |
 |
C610(0.00); C93(0.00); C69(0.00); C70(0.00) | LDD0236 | [3] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C121(0.51); C322(0.47) | LDD2142 | [5] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C121(1.33) | LDD2117 | [5] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C121(9.53); C322(4.80); C93(1.66) | LDD1702 | [5] |
| LDCM0625 | F8 | Ramos | C322(3.45); C185(1.34); 1.44; C610(0.88) | LDD2187 | [7] |
| LDCM0572 | Fragment10 | Ramos | C322(0.43); C185(0.28); 3.62; C610(0.52) | LDD2189 | [7] |
| LDCM0573 | Fragment11 | Ramos | C121(0.85); C322(0.72); C185(0.31); 9.76 | LDD2190 | [7] |
| LDCM0574 | Fragment12 | Ramos | C322(0.89); C610(0.59); C93(1.38) | LDD2191 | [7] |
| LDCM0575 | Fragment13 | Ramos | C322(0.47); 1.12; C610(0.91); C93(0.98) | LDD2192 | [7] |
| LDCM0576 | Fragment14 | Ramos | C121(1.04); C322(1.35); C185(0.46); 1.37 | LDD2193 | [7] |
| LDCM0579 | Fragment20 | Ramos | C610(0.61) | LDD2194 | [7] |
| LDCM0580 | Fragment21 | Ramos | 0.99; C610(0.60); C93(1.01) | LDD2195 | [7] |
| LDCM0582 | Fragment23 | Ramos | C322(0.57); 0.61; C610(0.87); C93(0.76) | LDD2196 | [7] |
| LDCM0578 | Fragment27 | Ramos | C185(0.78); 0.68; C610(0.87); C93(0.84) | LDD2197 | [7] |
| LDCM0586 | Fragment28 | Ramos | C121(2.41); C322(0.91); 0.90; C610(0.56) | LDD2198 | [7] |
| LDCM0588 | Fragment30 | Ramos | C322(0.62); C185(0.31); 0.94; C610(0.80) | LDD2199 | [7] |
| LDCM0589 | Fragment31 | Ramos | 1.07; C610(0.92); C93(0.89) | LDD2200 | [7] |
| LDCM0590 | Fragment32 | Ramos | C322(0.60); C185(0.23); C610(0.51); C93(1.59) | LDD2201 | [7] |
| LDCM0468 | Fragment33 | Ramos | C322(0.56); 1.06; C610(0.72); C93(0.96) | LDD2202 | [7] |
| LDCM0596 | Fragment38 | Ramos | C322(0.36); 0.68; C610(0.51); C93(0.90) | LDD2203 | [7] |
| LDCM0566 | Fragment4 | Ramos | C322(1.63); C185(0.99); C610(0.83); C93(2.74) | LDD2184 | [7] |
| LDCM0610 | Fragment52 | Ramos | C322(0.32); C93(1.24) | LDD2204 | [7] |
| LDCM0614 | Fragment56 | Ramos | C322(0.50); C185(0.32); 1.57; C610(0.66) | LDD2205 | [7] |
| LDCM0569 | Fragment7 | Ramos | C322(1.42); C185(1.02); 9.60; C610(0.66) | LDD2186 | [7] |
| LDCM0571 | Fragment9 | Ramos | C185(0.25); C610(0.72); C93(2.47) | LDD2188 | [7] |
| LDCM0015 | HNE | MDA-MB-231 | C322(1.02) | LDD0346 | [7] |
| LDCM0022 | KB02 | Ramos | C322(1.38); C185(2.29); C610(0.47); C93(1.44) | LDD2182 | [7] |
| LDCM0023 | KB03 | MDA-MB-231 | C121(9.77); C93(4.46); C185(1.13) | LDD1701 | [5] |
| LDCM0024 | KB05 | G361 | C177(2.54); C93(2.75); C666(1.87); C378(2.64) | LDD3311 | [4] |
| LDCM0509 | N-(4-bromo-3,5-dimethylphenyl)-2-nitroacetamide | MDA-MB-231 | C121(1.09) | LDD2102 | [5] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C121(1.15) | LDD2090 | [5] |
| LDCM0499 | Nucleophilic fragment 12b | MDA-MB-231 | C121(0.98) | LDD2092 | [5] |
| LDCM0501 | Nucleophilic fragment 13b | MDA-MB-231 | C121(1.12) | LDD2094 | [5] |
| LDCM0505 | Nucleophilic fragment 15b | MDA-MB-231 | C121(1.05) | LDD2098 | [5] |
| LDCM0506 | Nucleophilic fragment 16a | MDA-MB-231 | C121(1.09) | LDD2099 | [5] |
| LDCM0507 | Nucleophilic fragment 16b | MDA-MB-231 | C121(0.97) | LDD2100 | [5] |
| LDCM0511 | Nucleophilic fragment 18b | MDA-MB-231 | C121(0.68) | LDD2104 | [5] |
| LDCM0512 | Nucleophilic fragment 19a | MDA-MB-231 | C121(1.43) | LDD2105 | [5] |
| LDCM0513 | Nucleophilic fragment 19b | MDA-MB-231 | C121(0.56) | LDD2106 | [5] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C121(0.97) | LDD2107 | [5] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C121(1.48) | LDD2109 | [5] |
| LDCM0517 | Nucleophilic fragment 21b | MDA-MB-231 | C121(0.90) | LDD2110 | [5] |
| LDCM0526 | Nucleophilic fragment 26a | MDA-MB-231 | C121(1.67) | LDD2119 | [5] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C121(1.20) | LDD2123 | [5] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C121(1.10) | LDD2129 | [5] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C322(1.69) | LDD2134 | [5] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C121(1.12) | LDD2135 | [5] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C121(0.88) | LDD2140 | [5] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C121(0.87) | LDD2141 | [5] |
| LDCM0550 | Nucleophilic fragment 5a | MDA-MB-231 | C121(1.71) | LDD2144 | [5] |
| LDCM0551 | Nucleophilic fragment 5b | MDA-MB-231 | C121(1.03) | LDD2145 | [5] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C121(1.32) | LDD2146 | [5] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C121(2.10) | LDD2153 | [5] |
| LDCM0131 | RA190 | MM1.R | 2.07 | LDD0299 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Transporter and channel
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Reticulophagy regulator 3 (RETREG3) | RETREG family | Q86VR2 | |||
| Receptor-binding cancer antigen expressed on SiSo cells (EBAG9) | . | O00559 | |||
References
