Details of the Target
General Information of Target
| Target ID | LDTP12116 | |||||
|---|---|---|---|---|---|---|
| Target Name | Uridine-cytidine kinase 1 (UCK1) | |||||
| Gene Name | UCK1 | |||||
| Gene ID | 83549 | |||||
| Synonyms |
URK1; Uridine-cytidine kinase 1; UCK 1; EC 2.7.1.48; Cytidine monophosphokinase 1; Uridine monophosphokinase 1 |
|||||
| 3D Structure | ||||||
| Sequence |
MILLQHAVLPPPKQPSPSPPMSVATRSTGTLQLPPQKPFGQEASLPLAGEEELSKGGEQD
CALEELCKPLYCKLCNVTLNSAQQAQAHYQGKNHGKKLRNYYAANSCPPPARMSNVVEPA ATPVVPVPPQMGSFKPGGRVILATENDYCKLCDASFSSPAVAQAHYQGKNHAKRLRLAEA QSNSFSESSELGQRRARKEGNEFKMMPNRRNMYTVQNNSAGPYFNPRSRQRIPRDLAMCV TPSGQFYCSMCNVGAGEEMEFRQHLESKQHKSKVSEQRYRNEMENLGYV |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Uridine kinase family
|
|||||
| Function |
Phosphorylates uridine and cytidine to uridine monophosphate and cytidine monophosphate. Does not phosphorylate deoxyribonucleosides or purine ribonucleosides. Can use ATP or GTP as a phosphate donor. Can also phosphorylate cytidine and uridine nucleoside analogs such as 6-azauridine, 5-fluorouridine, 4-thiouridine, 5-bromouridine, N(4)-acetylcytidine, N(4)-benzoylcytidine, 5-fluorocytidine, 2-thiocytidine, 5-methylcytidine, and N(4)-anisoylcytidine.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
TH214 Probe Info |
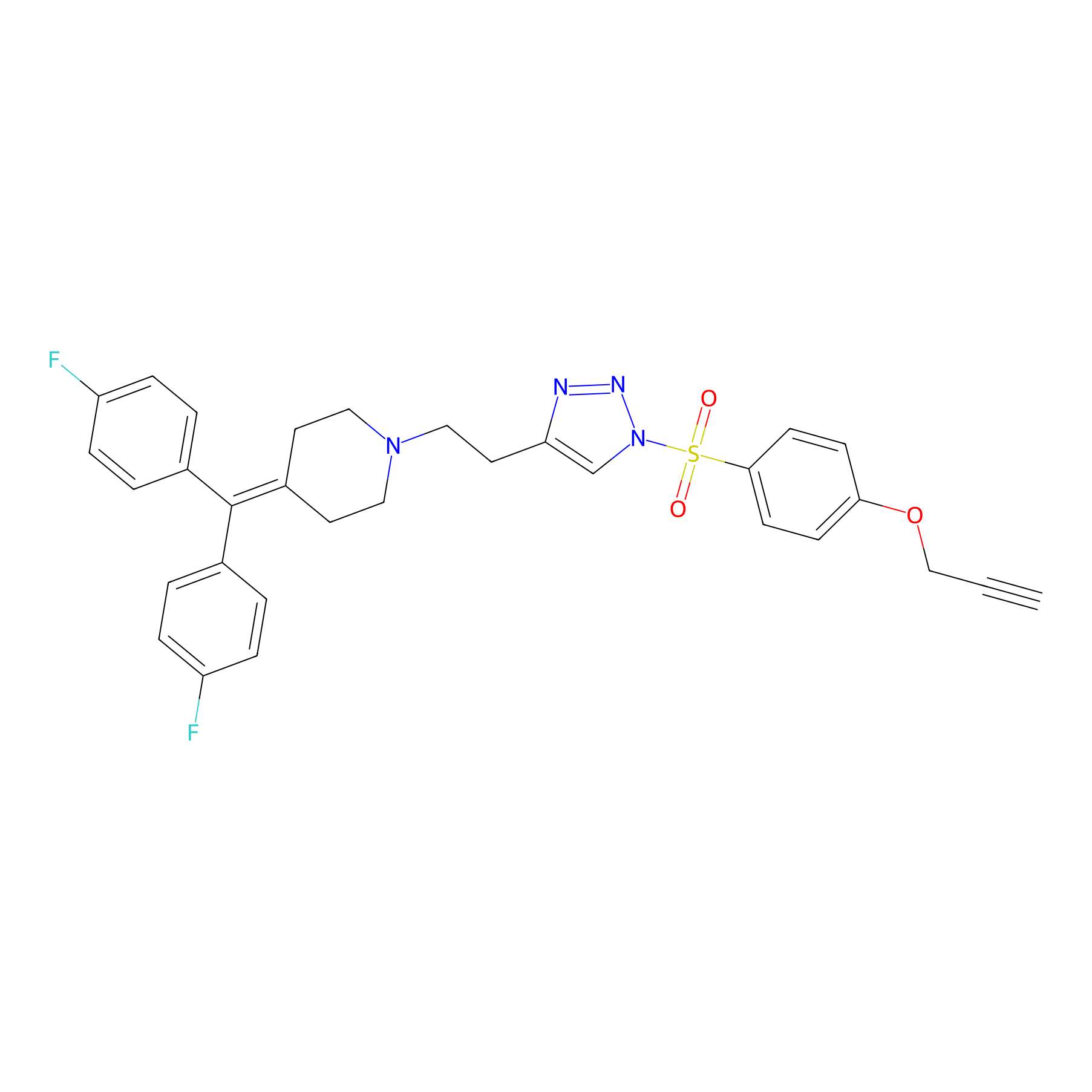 |
Y205(20.00) | LDD0258 | [2] | |
|
DBIA Probe Info |
 |
C241(1.18) | LDD3344 | [3] | |
|
EA-probe Probe Info |
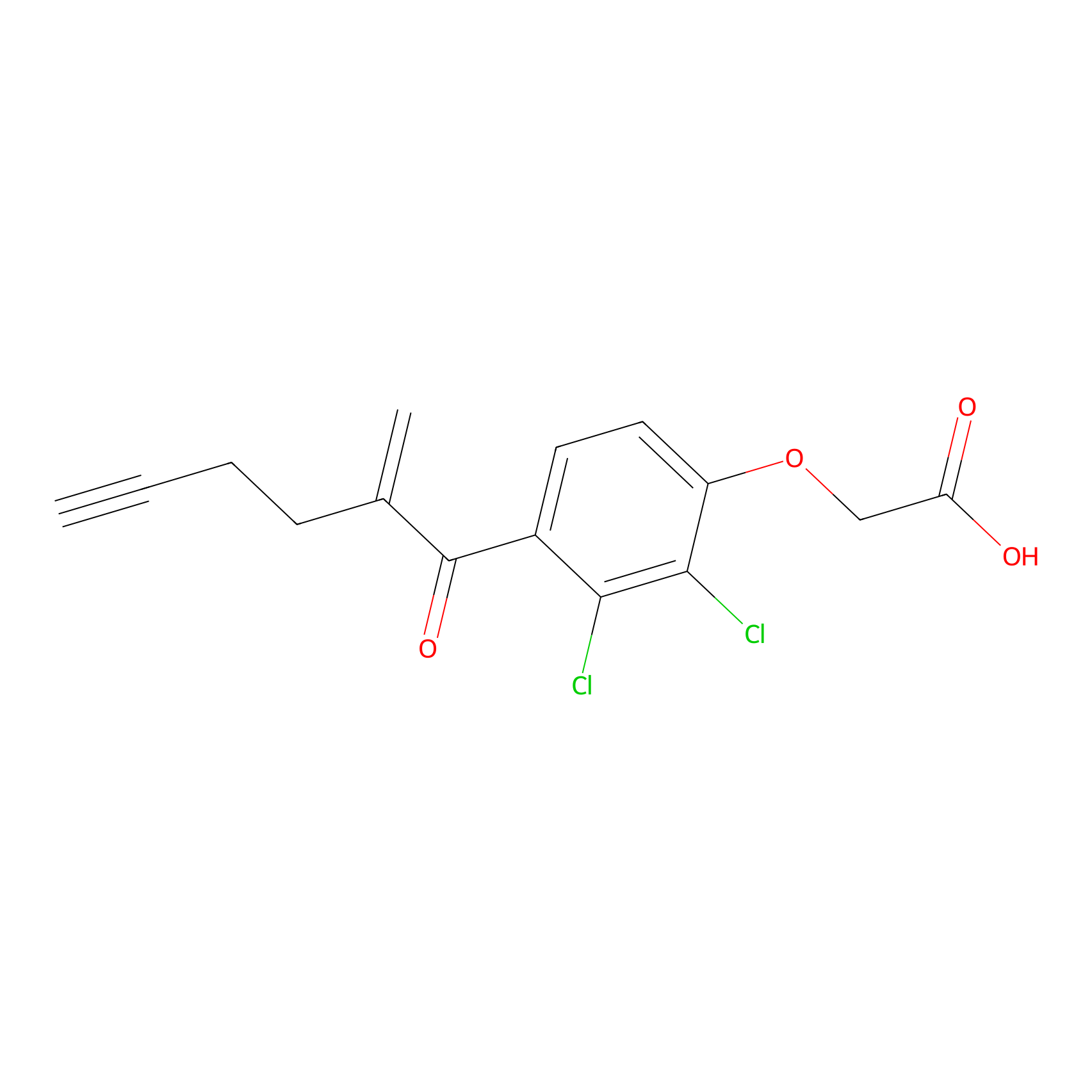 |
N.A. | LDD0440 | [4] | |
|
IPM Probe Info |
 |
C199(1.05) | LDD1701 | [5] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
N.A. | LDD0038 | [6] | |
|
IA-alkyne Probe Info |
 |
C199(0.00); C236(0.00) | LDD0036 | [6] | |
|
IPIAA_L Probe Info |
 |
N.A. | LDD0031 | [7] | |
|
Lodoacetamide azide Probe Info |
 |
C199(0.00); C236(0.00) | LDD0037 | [6] | |
|
AOyne Probe Info |
 |
15.00 | LDD0443 | [8] | |
|
NAIA_5 Probe Info |
 |
C199(0.00); C236(0.00) | LDD2223 | [9] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C236(2.35) | LDD0372 | [10] |
| LDCM0156 | Aniline | NCI-H1299 | 12.87 | LDD0403 | [1] |
| LDCM0175 | Ethacrynic acid | HeLa | N.A. | LDD0440 | [4] |
| LDCM0022 | KB02 | IGR-37 | C45(2.43) | LDD2396 | [3] |
| LDCM0023 | KB03 | MDA-MB-231 | C199(1.05) | LDD1701 | [5] |
| LDCM0024 | KB05 | NCI-H146 | C241(1.18) | LDD3344 | [3] |
The Interaction Atlas With This Target
References
