Details of the Target
General Information of Target
| Target ID | LDTP09996 | |||||
|---|---|---|---|---|---|---|
| Target Name | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1 (SMARCE1) | |||||
| Gene Name | SMARCE1 | |||||
| Gene ID | 6605 | |||||
| Synonyms |
BAF57; SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily E member 1; BRG1-associated factor 57; BAF57 |
|||||
| 3D Structure | ||||||
| Sequence |
MMQSATVPAEGAVKGLPEMLGVPMQQIPQCAGCNQHILDKFILKVLDRHWHSSCLKCADC
QMQLADRCFSRAGSVYCKEDFFKRFGTKCTACQQGIPPTQVVRKAQDFVYHLHCFACIIC NRQLATGDEFYLMEDGRLVCKEDYETAKQNDDSEAGAKRPRTTITAKQLETLKNAYKNSP KPARHVREQLSSETGLDMRVVQVWFQNRRAKEKRLKKDAGRHRWGQFYKSVKRSRGSSKQ EKESSAEDCGVSDSELSFREDQILSELGHTNRIYGNVGDVTGGQLMNGSFSMDGTGQSYQ DLRDGSPYGIPQSPSSISSLPSHAPLLNGLDYTVDSNLGIIAHAGQGVSQTLRAMAGGPT SDISTGSSVGYPDFPTSPGSWLDEMDHPPF |
|||||
| Target Bioclass |
Transcription factor
|
|||||
| Subcellular location |
Nucleus
|
|||||
| Function |
Involved in transcriptional activation and repression of select genes by chromatin remodeling (alteration of DNA-nucleosome topology). Component of SWI/SNF chromatin remodeling complexes that carry out key enzymatic activities, changing chromatin structure by altering DNA-histone contacts within a nucleosome in an ATP-dependent manner. Belongs to the neural progenitors-specific chromatin remodeling complex (npBAF complex) and the neuron-specific chromatin remodeling complex (nBAF complex). During neural development a switch from a stem/progenitor to a postmitotic chromatin remodeling mechanism occurs as neurons exit the cell cycle and become committed to their adult state. The transition from proliferating neural stem/progenitor cells to postmitotic neurons requires a switch in subunit composition of the npBAF and nBAF complexes. As neural progenitors exit mitosis and differentiate into neurons, npBAF complexes which contain ACTL6A/BAF53A and PHF10/BAF45A, are exchanged for homologous alternative ACTL6B/BAF53B and DPF1/BAF45B or DPF3/BAF45C subunits in neuron-specific complexes (nBAF). The npBAF complex is essential for the self-renewal/proliferative capacity of the multipotent neural stem cells. The nBAF complex along with CREST plays a role regulating the activity of genes essential for dendrite growth. Required for the coactivation of estrogen responsive promoters by SWI/SNF complexes and the SRC/p160 family of histone acetyltransferases (HATs). Also specifically interacts with the CoREST corepressor resulting in repression of neuronal specific gene promoters in non-neuronal cells.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
AZ-9 Probe Info |
 |
10.00 | LDD0393 | [2] | |
|
C-Sul Probe Info |
 |
5.75 | LDD0066 | [3] | |
|
FBP2 Probe Info |
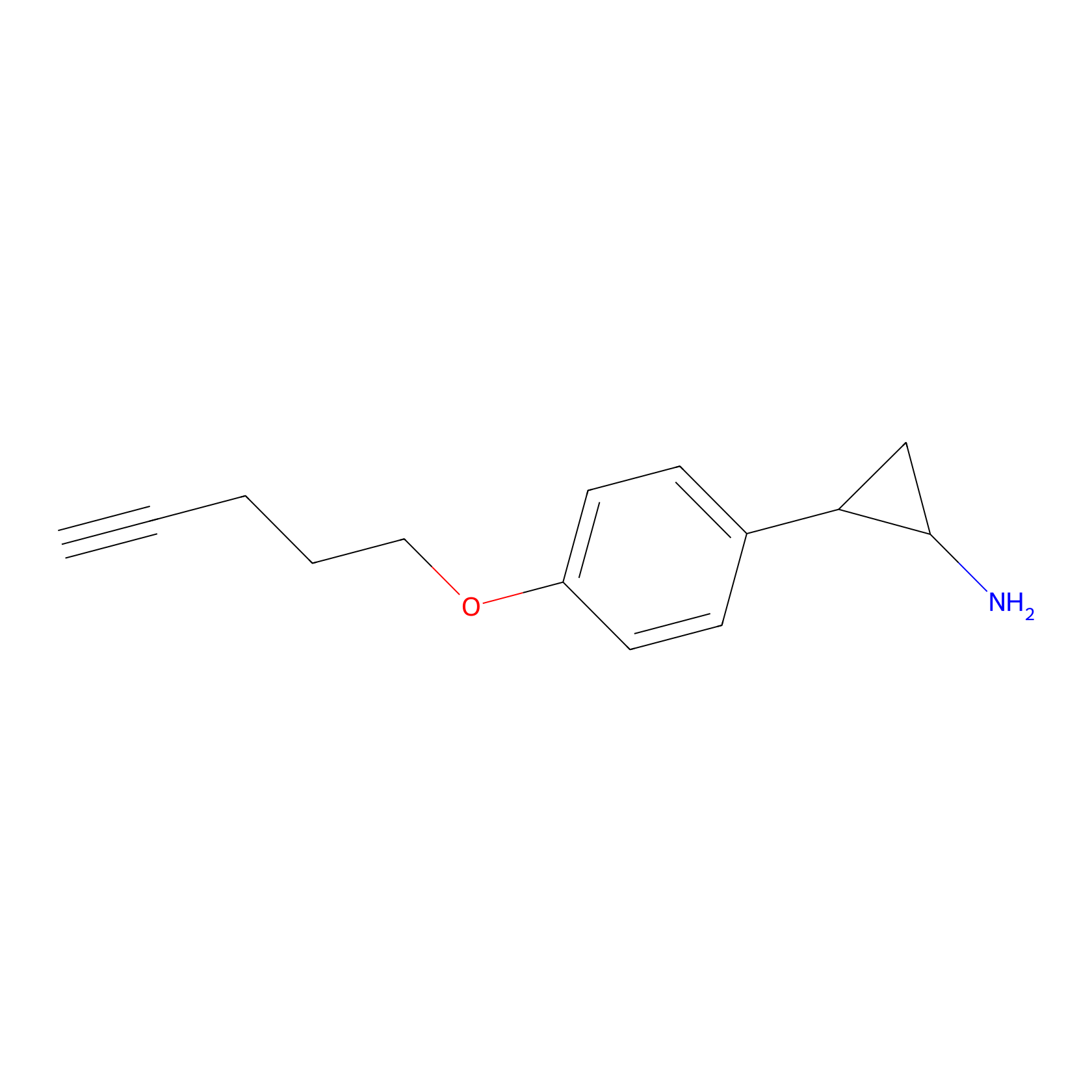 |
2.14 | LDD0323 | [4] | |
|
TH211 Probe Info |
 |
Y119(20.00); Y36(20.00) | LDD0257 | [5] | |
|
Probe 1 Probe Info |
 |
Y115(13.26); Y119(15.23) | LDD3495 | [6] | |
|
BTD Probe Info |
 |
C274(0.67) | LDD2108 | [7] | |
|
HHS-475 Probe Info |
 |
Y142(0.76); Y31(0.86); Y119(1.87) | LDD0264 | [8] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0224 | [9] | |
|
5E-2FA Probe Info |
 |
N.A. | LDD2235 | [10] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [11] | |
|
Lodoacetamide azide Probe Info |
 |
N.A. | LDD0037 | [11] | |
|
SF Probe Info |
 |
N.A. | LDD0028 | [12] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [13] | |
|
1c-yne Probe Info |
 |
N.A. | LDD0228 | [14] | |
|
HHS-465 Probe Info |
 |
K123(0.00); Y119(0.00) | LDD2240 | [15] | |
|
HHS-482 Probe Info |
 |
Y119(0.99); Y36(1.33) | LDD2239 | [16] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0548 | 1-(4-(Benzo[d][1,3]dioxol-5-ylmethyl)piperazin-1-yl)-2-nitroethan-1-one | MDA-MB-231 | C274(0.49) | LDD2142 | [7] |
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C274(0.60) | LDD2117 | [7] |
| LDCM0156 | Aniline | NCI-H1299 | 11.65 | LDD0403 | [1] |
| LDCM0116 | HHS-0101 | DM93 | Y142(0.76); Y31(0.86); Y119(1.87) | LDD0264 | [8] |
| LDCM0117 | HHS-0201 | DM93 | Y142(0.34); Y119(3.21) | LDD0265 | [8] |
| LDCM0118 | HHS-0301 | DM93 | Y142(0.81); Y119(2.88) | LDD0266 | [8] |
| LDCM0119 | HHS-0401 | DM93 | Y142(0.75); Y119(1.89) | LDD0267 | [8] |
| LDCM0120 | HHS-0701 | DM93 | Y142(1.18); Y31(2.34); Y119(3.56) | LDD0268 | [8] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0224 | [9] |
| LDCM0515 | Nucleophilic fragment 20b | MDA-MB-231 | C274(0.67) | LDD2108 | [7] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C274(0.53) | LDD2109 | [7] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C274(0.96) | LDD2111 | [7] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C274(1.09) | LDD2135 | [7] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C274(0.52) | LDD2140 | [7] |
| LDCM0547 | Nucleophilic fragment 41 | MDA-MB-231 | C274(0.55) | LDD2141 | [7] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Transporter and channel
Transcription factor
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| REST corepressor 1 (RCOR1) | CoREST family | Q9UKL0 | |||
| AT-rich interactive domain-containing protein 2 (ARID2) | . | Q68CP9 | |||
| Homeobox protein MOX-2 (MEOX2) | . | P50222 | |||
Other
References
