Details of the Target
General Information of Target
| Target ID | LDTP08418 | |||||
|---|---|---|---|---|---|---|
| Target Name | RAS protein activator like-3 (RASAL3) | |||||
| Gene Name | RASAL3 | |||||
| Gene ID | 64926 | |||||
| Synonyms |
RAS protein activator like-3 |
|||||
| 3D Structure | ||||||
| Sequence |
MDPPSPSRTSQTQPTATSPLTSYRWHTGGGGEKAAGGFRWGRFAGWGRALSHQEPMVSTQ
PAPRSIFRRVLSAPPKESRTSRLRLSKALWGRHKNPPPEPDPEPEQEAPELEPEPELEPP TPQIPEAPTPNVPVWDIGGFTLLDGKLVLLGGEEEGPRRPRVGSASSEGSIHVAMGNFRD PDRMPGKTEPETAGPNQVHNVRGLLKRLKEKKKARLEPRDGPPSALGSRESLATLSELDL GAERDVRIWPLHPSLLGEPHCFQVTWTGGSRCFSCRSAAERDRWIEDLRRQFQPTQDNVE REETWLSVWVHEAKGLPRAAAGAPGVRAELWLDGALLARTAPRAGPGQLFWAERFHFEAL PPARRLSLRLRGLGPGSAVLGRVALALEELDAPRAPAAGLERWFPLLGAPAGAALRARIR ARRLRVLPSERYKELAEFLTFHYARLCGALEPALPAQAKEELAAAMVRVLRATGRAQALV TDLGTAELARCGGREALLFRENTLATKAIDEYMKLVAQDYLQETLGQVVRRLCASTEDCE VDPSKCPASELPEHQARLRNSCEEVFETIIHSYDWFPAELGIVFSSWREACKERGSEVLG PRLVCASLFLRLLCPAILAPSLFGLAPDHPAPGPARTLTLIAKVIQNLANRAPFGEKEAY MGFMNSFLEEHGPAMQCFLDQVAMVDVDAAPSGYQGSGDLALQLAVLHAQLCTIFAELDQ TTRDTLEPLPTILRAIEEGQPVLVSVPMRLPLPPAQVHSSLSAGEKPGFLAPRDLPKHTP LISKSQSLRSVRRSESWARPRPDEERPLRRPRPVQRTQSVPVRRPARRRQSAGPWPRPKG SLSMGPAPRARPWTRDSASLPRKPSVPWQRQMDQPQDRNQALGTHRPVNKLAELQCEVAA LREEQKVLSRLVESLSTQIRALTEQQEQLRGQLQDLDSRLRAGSSEFDSEHNLTSNEGHS LKNLEHRLNEMERTQAQLRDAVQSLQLSPRTRGSWSQPQPLKAPCLNGDTT |
|||||
| Target Bioclass |
Other
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
Functions as a Ras GTPase-activating protein. Plays an important role in the expansion and functions of natural killer T (NKT) cells in the liver by negatively regulating RAS activity and the down-stream ERK signaling pathway.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| BXPC3 | SNV: p.R812W | . | |||
| HCT116 | SNV: p.Q876H | . | |||
| HCT15 | SNV: p.R327C | . | |||
| IGR1 | SNV: p.D873A | . | |||
| IM95 | Deletion: p.P120LfsTer89 | . | |||
| JURKAT | SNV: p.K33N | Compound 10 Probe Info | |||
| MCC26 | Substitution: p.P122S | . | |||
| MOLT4 | SNV: p.R490H | IA-alkyne Probe Info | |||
| OVCAR5 | SNV: p.E795D | . | |||
| REH | SNV: p.R886Q | DBIA Probe Info | |||
| SBC5 | Deletion: p.R382AfsTer99 | . | |||
| SKMEL2 | SNV: p.A62T | . | |||
| SKOV3 | SNV: p.R215W | . | |||
| SNU1 | SNV: p.L205I | . | |||
| TE4 | SNV: p.E727V | . | |||
| TOV21G | Substitution: p.Q876K | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
HPAP Probe Info |
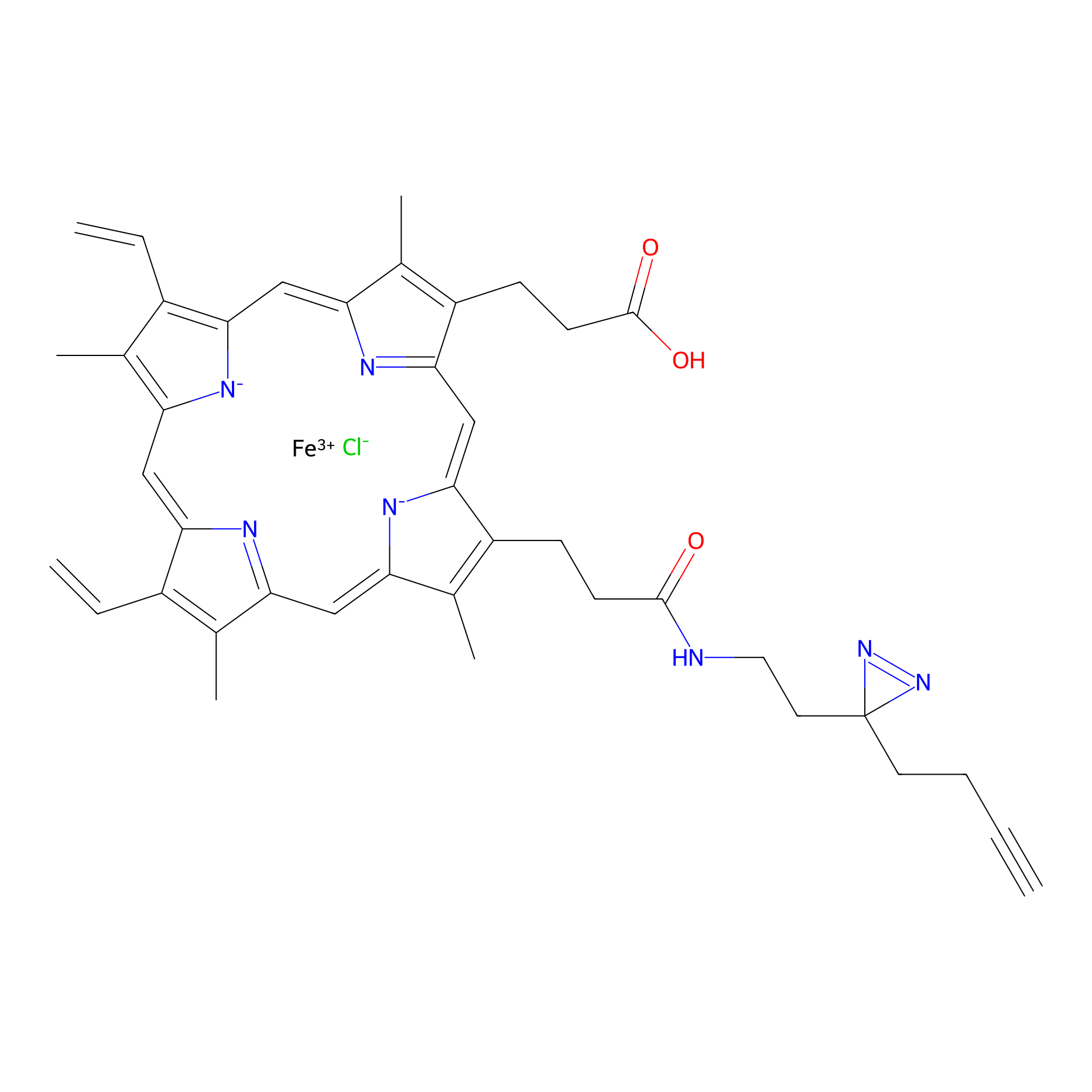 |
6.67 | LDD0064 | [1] | |
|
Alkyne-RA190 Probe Info |
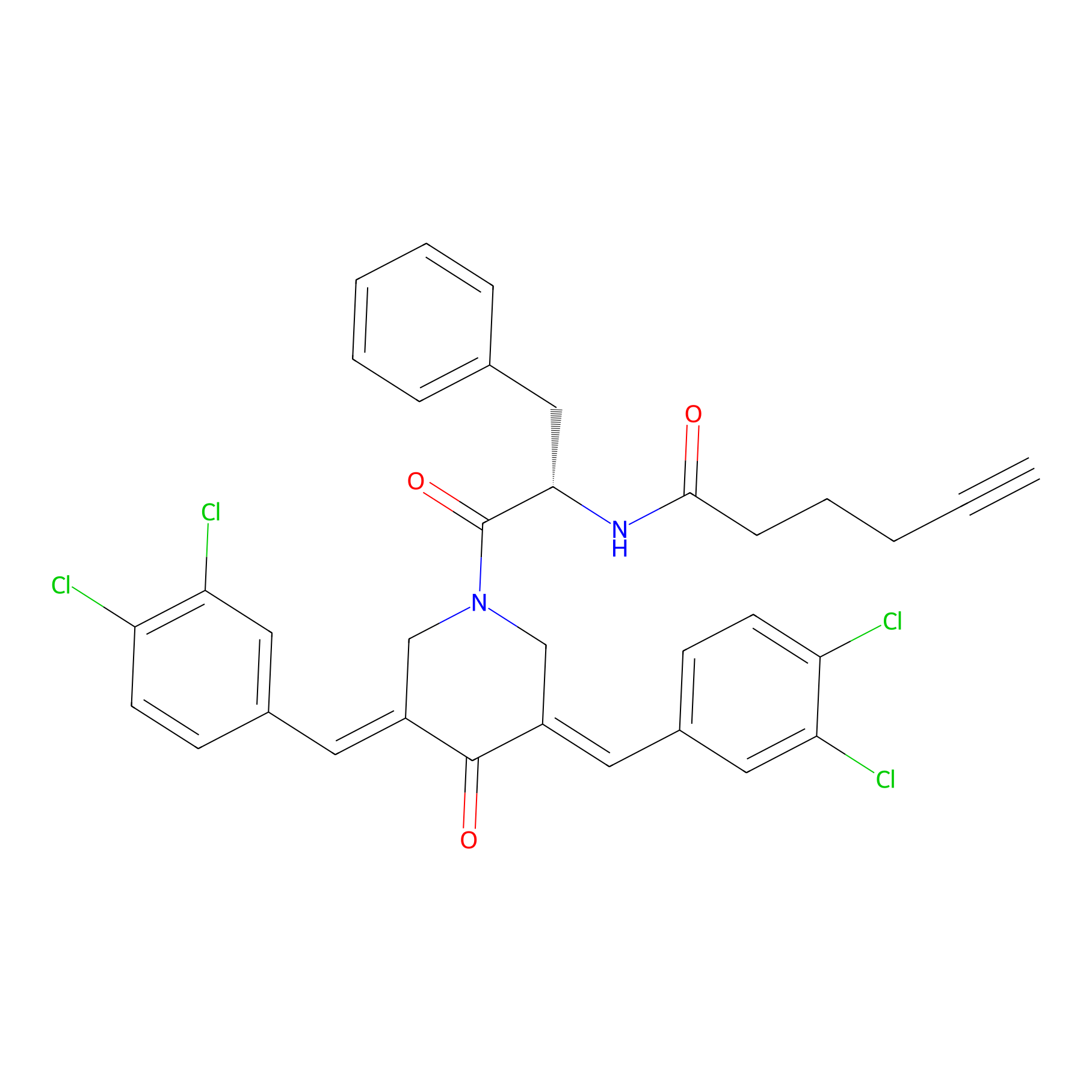 |
3.33 | LDD0300 | [2] | |
|
DBIA Probe Info |
 |
C605(48.74); C546(44.85) | LDD0209 | [3] | |
|
4-Iodoacetamidophenylacetylene Probe Info |
 |
C896(0.00); C614(0.00); C546(0.00); C605(0.00) | LDD0038 | [4] | |
|
IA-alkyne Probe Info |
 |
C447(0.00); C614(0.00); C605(0.00); C896(0.00) | LDD0036 | [4] | |
|
Lodoacetamide azide Probe Info |
 |
C546(0.00); C896(0.00); C447(0.00); C614(0.00) | LDD0037 | [4] | |
|
Compound 10 Probe Info |
 |
N.A. | LDD2216 | [5] | |
|
Compound 11 Probe Info |
 |
N.A. | LDD2213 | [5] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0625 | F8 | Ramos | C896(1.64); C1005(1.42) | LDD2187 | [6] |
| LDCM0572 | Fragment10 | Ramos | C896(0.53); C1005(1.02) | LDD2189 | [6] |
| LDCM0573 | Fragment11 | Ramos | C896(2.49) | LDD2190 | [6] |
| LDCM0574 | Fragment12 | Ramos | C896(0.65); C1005(0.50) | LDD2191 | [6] |
| LDCM0575 | Fragment13 | Ramos | C1005(0.63) | LDD2192 | [6] |
| LDCM0576 | Fragment14 | Ramos | C1005(0.98) | LDD2193 | [6] |
| LDCM0579 | Fragment20 | Ramos | C896(0.65); C1005(0.43) | LDD2194 | [6] |
| LDCM0580 | Fragment21 | Ramos | C1005(0.74) | LDD2195 | [6] |
| LDCM0582 | Fragment23 | Ramos | C1005(0.45) | LDD2196 | [6] |
| LDCM0586 | Fragment28 | Ramos | C896(1.70); C1005(0.48) | LDD2198 | [6] |
| LDCM0588 | Fragment30 | Ramos | C896(1.03); C1005(0.79) | LDD2199 | [6] |
| LDCM0590 | Fragment32 | Ramos | C896(0.90) | LDD2201 | [6] |
| LDCM0468 | Fragment33 | Ramos | C896(1.13) | LDD2202 | [6] |
| LDCM0596 | Fragment38 | Ramos | C896(1.61); C1005(0.32) | LDD2203 | [6] |
| LDCM0610 | Fragment52 | Ramos | C896(0.46); C1005(0.62) | LDD2204 | [6] |
| LDCM0569 | Fragment7 | Ramos | C1005(0.99) | LDD2186 | [6] |
| LDCM0571 | Fragment9 | Ramos | C896(0.77) | LDD2188 | [6] |
| LDCM0022 | KB02 | 697 | C546(2.06) | LDD2245 | [7] |
| LDCM0023 | KB03 | Jurkat | C605(48.74); C546(44.85) | LDD0209 | [3] |
| LDCM0024 | KB05 | MOLM-13 | C546(3.59) | LDD3333 | [7] |
| LDCM0014 | Panhematin | hPBMC | 6.67 | LDD0064 | [1] |
| LDCM0131 | RA190 | MM1.R | 3.33 | LDD0300 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
References
