Details of the Target
General Information of Target
| Target ID | LDTP06482 | |||||
|---|---|---|---|---|---|---|
| Target Name | Ubiquitin-conjugating enzyme E2 variant 2 (UBE2V2) | |||||
| Gene Name | UBE2V2 | |||||
| Gene ID | 7336 | |||||
| Synonyms |
MMS2; UEV2; Ubiquitin-conjugating enzyme E2 variant 2; DDVit 1; Enterocyte differentiation-associated factor 1; EDAF-1; Enterocyte differentiation-promoting factor 1; EDPF-1; MMS2 homolog; Vitamin D3-inducible protein
|
|||||
| 3D Structure | ||||||
| Sequence |
MAVSTGVKVPRNFRLLEELEEGQKGVGDGTVSWGLEDDEDMTLTRWTGMIIGPPRTNYEN
RIYSLKVECGPKYPEAPPSVRFVTKINMNGINNSSGMVDARSIPVLAKWQNSYSIKVVLQ ELRRLMMSKENMKLPQPPEGQTYNN |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Ubiquitin-conjugating enzyme family
|
|||||
| Function |
Has no ubiquitin ligase activity on its own. The UBE2V2/UBE2N heterodimer catalyzes the synthesis of non-canonical poly-ubiquitin chains that are linked through 'Lys-63'. This type of poly-ubiquitination does not lead to protein degradation by the proteasome. Mediates transcriptional activation of target genes. Plays a role in the control of progress through the cell cycle and differentiation. Plays a role in the error-free DNA repair pathway and contributes to the survival of cells after DNA damage.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
m-APA Probe Info |
 |
15.00 | LDD0402 | [1] | |
|
FBP2 Probe Info |
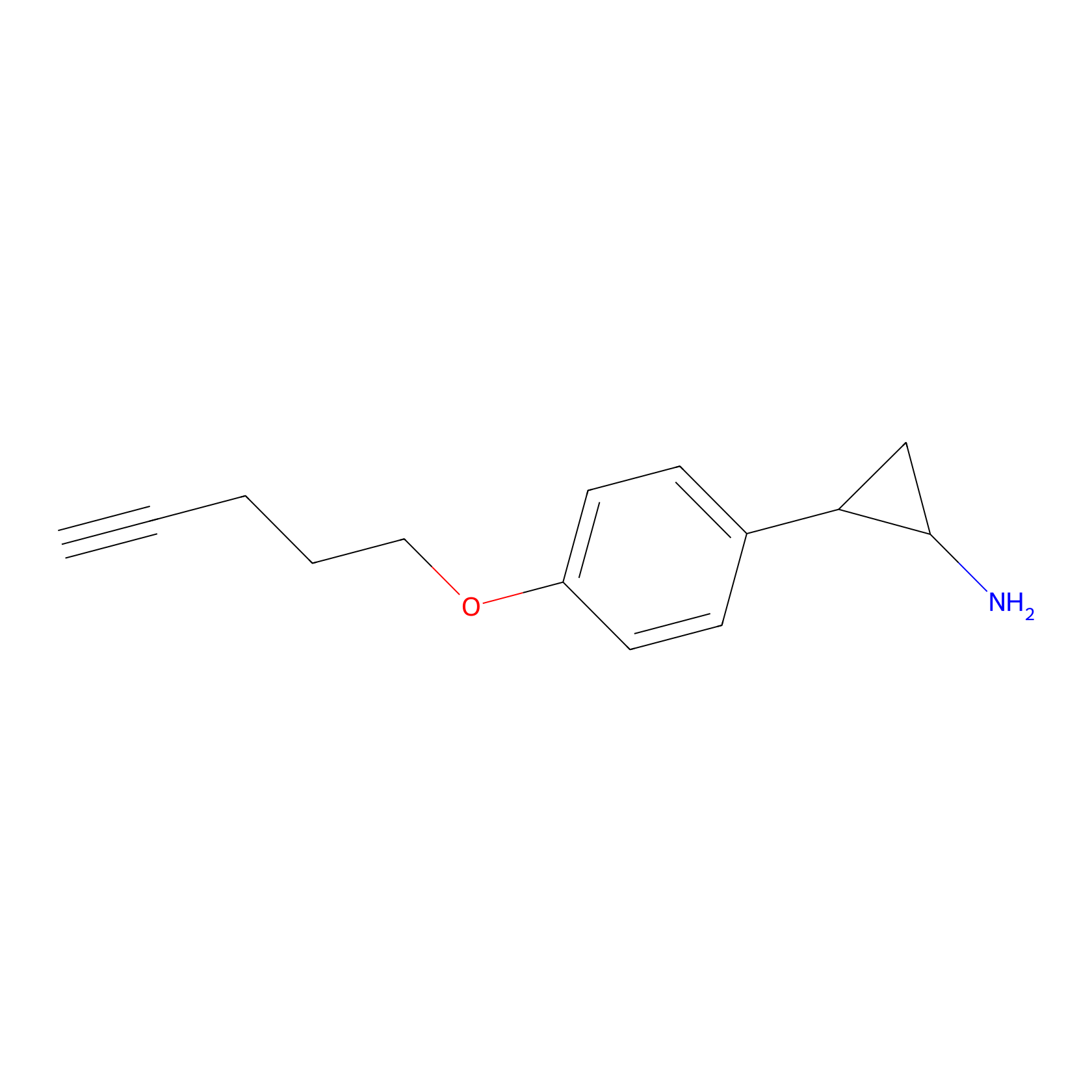 |
2.72 | LDD0323 | [2] | |
|
AZ-9 Probe Info |
 |
E20(10.00); E17(10.00) | LDD2209 | [3] | |
|
DA-P3 Probe Info |
 |
4.16 | LDD0183 | [4] | |
|
HHS-475 Probe Info |
 |
Y113(0.92) | LDD0264 | [5] | |
|
HHS-465 Probe Info |
 |
Y113(10.00) | LDD2237 | [6] | |
|
ATP probe Probe Info |
 |
K72(0.00); K66(0.00) | LDD0199 | [7] | |
|
IA-alkyne Probe Info |
 |
N.A. | LDD0036 | [8] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [9] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [10] | |
|
NHS Probe Info |
 |
K8(0.00); K129(0.00) | LDD0010 | [11] | |
|
1d-yne Probe Info |
 |
N.A. | LDD0357 | [12] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [13] | |
|
Crotonaldehyde Probe Info |
 |
N.A. | LDD0219 | [13] | |
|
Methacrolein Probe Info |
 |
N.A. | LDD0218 | [13] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [14] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0108 | Chloroacetamide | HeLa | N.A. | LDD0222 | [13] |
| LDCM0632 | CL-Sc | Hep-G2 | C69(20.00) | LDD2227 | [14] |
| LDCM0031 | Epigallocatechin gallate | HEK-293T | 4.16 | LDD0183 | [4] |
| LDCM0572 | Fragment10 | Ramos | C69(0.47) | LDD2189 | [15] |
| LDCM0573 | Fragment11 | Ramos | C69(1.48) | LDD2190 | [15] |
| LDCM0574 | Fragment12 | Ramos | C69(0.21) | LDD2191 | [15] |
| LDCM0575 | Fragment13 | Ramos | C69(0.58) | LDD2192 | [15] |
| LDCM0576 | Fragment14 | Ramos | C69(0.51) | LDD2193 | [15] |
| LDCM0579 | Fragment20 | Ramos | C69(0.19) | LDD2194 | [15] |
| LDCM0580 | Fragment21 | Ramos | C69(0.72) | LDD2195 | [15] |
| LDCM0582 | Fragment23 | Ramos | C69(3.37) | LDD2196 | [15] |
| LDCM0578 | Fragment27 | Ramos | C69(0.73) | LDD2197 | [15] |
| LDCM0588 | Fragment30 | Ramos | C69(0.54) | LDD2199 | [15] |
| LDCM0589 | Fragment31 | Ramos | C69(0.72) | LDD2200 | [15] |
| LDCM0590 | Fragment32 | Ramos | C69(0.34) | LDD2201 | [15] |
| LDCM0468 | Fragment33 | Ramos | C69(0.62) | LDD2202 | [15] |
| LDCM0596 | Fragment38 | Ramos | C69(0.71) | LDD2203 | [15] |
| LDCM0610 | Fragment52 | Ramos | C69(0.85) | LDD2204 | [15] |
| LDCM0571 | Fragment9 | Ramos | C69(0.34) | LDD2188 | [15] |
| LDCM0116 | HHS-0101 | DM93 | Y113(0.92) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y113(0.85) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y113(0.89) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y113(0.96) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y113(0.95) | LDD0268 | [5] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [13] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [13] |
The Interaction Atlas With This Target
References
