Details of the Target
General Information of Target
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| A549 | Deletion: p.R51AfsTer51 | . | |||
| HGC27 | SNV: p.E130K | . | |||
| JURKAT | SNV: p.P129H | . | |||
| L428 | SNV: p.C99R; p.S104T | . | |||
| LNCaP clone FGC | SNV: p.K306N | . | |||
| NCIH146 | Substitution: p.H278N | . | |||
| OCILY3 | SNV: p.A100T | DBIA Probe Info | |||
| P31FUJ | SNV: p.L378M | . | |||
| TOV21G | SNV: p.I118N | . | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K327(3.94) | LDD2218 | [1] | |
|
IA-alkyne Probe Info |
 |
C194(7.46); C19(1.99); C250(1.18); C343(1.05) | LDD0304 | [2] | |
|
DBIA Probe Info |
 |
C274(1.66); C343(1.31) | LDD3332 | [3] | |
|
Alkyne-RA190 Probe Info |
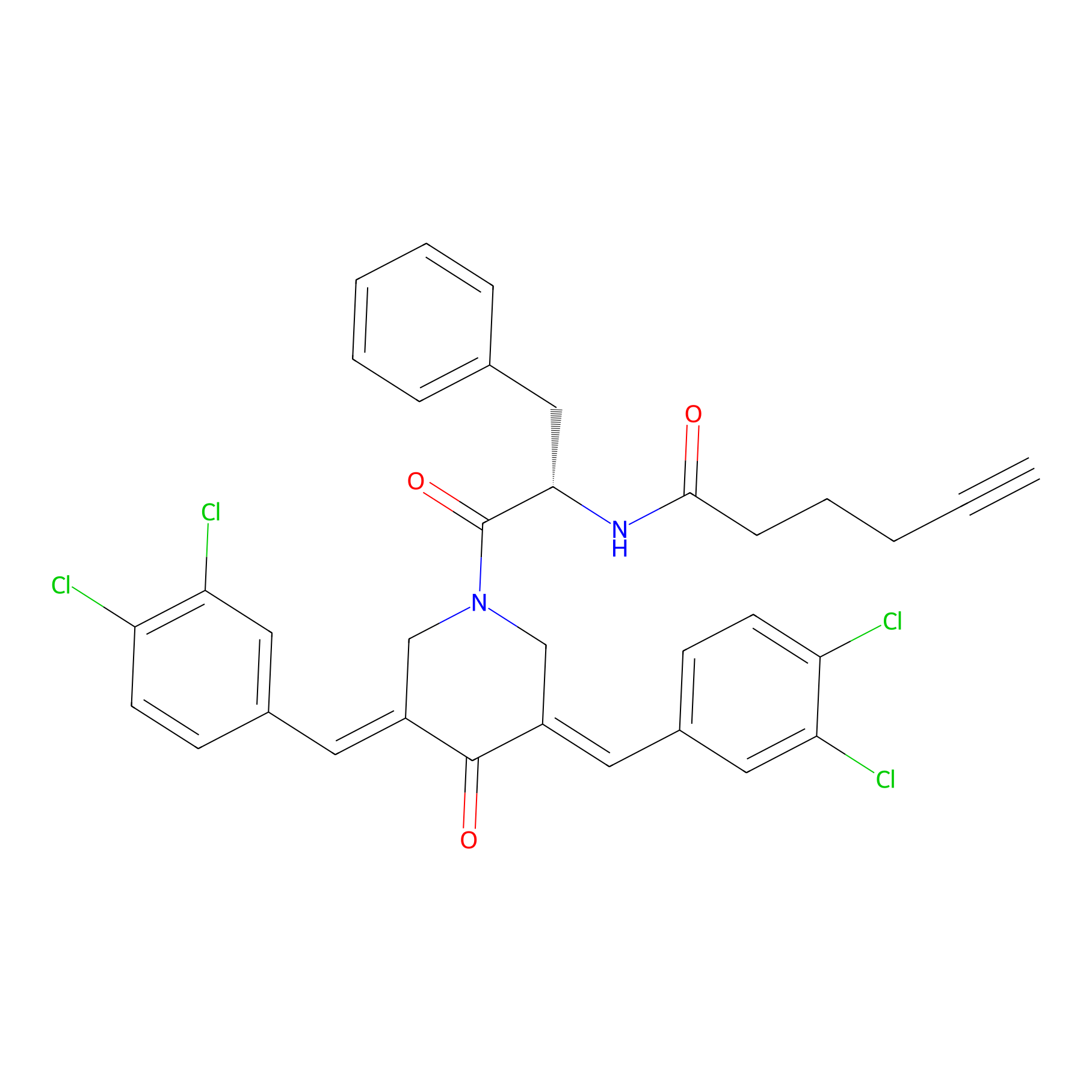 |
2.80 | LDD0299 | [2] | |
|
HHS-475 Probe Info |
 |
Y428(0.05); Y415(1.71) | LDD0264 | [4] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0625 | F8 | Ramos | C250(1.21); C19(1.34); C194(1.15); C343(1.48) | LDD2187 | [5] |
| LDCM0572 | Fragment10 | Ramos | C250(0.34); C19(0.99); C194(3.67); C343(0.40) | LDD2189 | [5] |
| LDCM0573 | Fragment11 | Ramos | C250(0.25); C19(0.38); C194(0.44); C343(0.34) | LDD2190 | [5] |
| LDCM0574 | Fragment12 | Ramos | C250(0.63); C19(1.50); C194(2.68); C343(0.30) | LDD2191 | [5] |
| LDCM0575 | Fragment13 | Ramos | C250(0.84); C19(1.20); C194(1.12); C343(0.57) | LDD2192 | [5] |
| LDCM0576 | Fragment14 | Ramos | C250(0.73); C19(0.81); C194(0.98); C343(0.68) | LDD2193 | [5] |
| LDCM0579 | Fragment20 | Ramos | C250(0.73); C19(1.10); C194(3.51); C343(0.30) | LDD2194 | [5] |
| LDCM0580 | Fragment21 | Ramos | C250(0.80); C19(0.95); C194(1.03); C343(0.36) | LDD2195 | [5] |
| LDCM0582 | Fragment23 | Ramos | C250(0.44); C19(1.31); C194(0.89); C343(0.68) | LDD2196 | [5] |
| LDCM0578 | Fragment27 | Ramos | C250(0.95); C19(0.82); C194(0.92); C343(1.07) | LDD2197 | [5] |
| LDCM0586 | Fragment28 | Ramos | C250(1.42); C19(0.62); C194(0.67); C343(0.53) | LDD2198 | [5] |
| LDCM0588 | Fragment30 | Ramos | C250(0.75); C19(1.41); C194(1.31); C343(0.52) | LDD2199 | [5] |
| LDCM0589 | Fragment31 | Ramos | C250(0.85); C19(1.10); C194(1.03); C343(0.64) | LDD2200 | [5] |
| LDCM0590 | Fragment32 | Ramos | C250(0.52); C19(1.21); C194(3.19); C343(0.34) | LDD2201 | [5] |
| LDCM0468 | Fragment33 | Ramos | C250(0.68); C19(1.28); C194(1.39); C343(0.41) | LDD2202 | [5] |
| LDCM0596 | Fragment38 | Ramos | C250(0.76); C19(1.39); C194(0.92); C343(0.63) | LDD2203 | [5] |
| LDCM0566 | Fragment4 | Ramos | C250(0.90); C19(0.77); C194(2.30); C343(0.72) | LDD2184 | [5] |
| LDCM0610 | Fragment52 | Ramos | C250(0.70); C19(1.33); C194(1.14); C343(0.75) | LDD2204 | [5] |
| LDCM0614 | Fragment56 | Ramos | C250(0.88); C19(1.26); C194(0.94); C343(0.50) | LDD2205 | [5] |
| LDCM0569 | Fragment7 | Ramos | C250(0.66); C19(0.77); C194(1.96); C343(0.64) | LDD2186 | [5] |
| LDCM0571 | Fragment9 | Ramos | C250(0.38); C19(1.50); C194(3.20); C343(0.32) | LDD2188 | [5] |
| LDCM0116 | HHS-0101 | DM93 | Y428(0.05); Y415(1.71) | LDD0264 | [4] |
| LDCM0117 | HHS-0201 | DM93 | Y415(1.23) | LDD0265 | [4] |
| LDCM0118 | HHS-0301 | DM93 | Y415(1.46) | LDD0266 | [4] |
| LDCM0119 | HHS-0401 | DM93 | Y415(0.73) | LDD0267 | [4] |
| LDCM0120 | HHS-0701 | DM93 | Y428(0.05); Y415(1.80) | LDD0268 | [4] |
| LDCM0022 | KB02 | Ramos | 6.24 | LDD0431 | [6] |
| LDCM0023 | KB03 | Ramos | C250(0.77); C19(0.86); C194(1.43); C343(0.67) | LDD2183 | [5] |
| LDCM0024 | KB05 | MKN45 | C274(1.66); C343(1.31) | LDD3332 | [3] |
| LDCM0131 | RA190 | MM1.R | 2.80 | LDD0299 | [2] |
| LDCM0170 | Structure8 | Ramos | 7.13; 4.52 | LDD0433 | [6] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Elongator complex protein 1 (ELP1) | ELP1/IKA1 family | O95163 | |||
| PDZ domain-containing protein GIPC2 (GIPC2) | GIPC family | Q8TF65 | |||
References
