Details of the Target
General Information of Target
| Target ID | LDTP05554 | |||||
|---|---|---|---|---|---|---|
| Target Name | Epithelial discoidin domain-containing receptor 1 (DDR1) | |||||
| Gene Name | DDR1 | |||||
| Gene ID | 780 | |||||
| Synonyms |
CAK; EDDR1; NEP; NTRK4; PTK3A; RTK6; TRKE; Epithelial discoidin domain-containing receptor 1; Epithelial discoidin domain receptor 1; EC 2.7.10.1; CD167 antigen-like family member A; Cell adhesion kinase; Discoidin receptor tyrosine kinase; HGK2; Mammary carcinoma kinase 10; MCK-10; Protein-tyrosine kinase 3A; Protein-tyrosine kinase RTK-6; TRK E; Tyrosine kinase DDR; Tyrosine-protein kinase CAK; CD antigen CD167a
|
|||||
| 3D Structure | ||||||
| Sequence |
MGPEALSSLLLLLLVASGDADMKGHFDPAKCRYALGMQDRTIPDSDISASSSWSDSTAAR
HSRLESSDGDGAWCPAGSVFPKEEEYLQVDLQRLHLVALVGTQGRHAGGLGKEFSRSYRL RYSRDGRRWMGWKDRWGQEVISGNEDPEGVVLKDLGPPMVARLVRFYPRADRVMSVCLRV ELYGCLWRDGLLSYTAPVGQTMYLSEAVYLNDSTYDGHTVGGLQYGGLGQLADGVVGLDD FRKSQELRVWPGYDYVGWSNHSFSSGYVEMEFEFDRLRAFQAMQVHCNNMHTLGARLPGG VECRFRRGPAMAWEGEPMRHNLGGNLGDPRARAVSVPLGGRVARFLQCRFLFAGPWLLFS EISFISDVVNNSSPALGGTFPPAPWWPPGPPPTNFSSLELEPRGQQPVAKAEGSPTAILI GCLVAIILLLLLIIALMLWRLHWRRLLSKAERRVLEEELTVHLSVPGDTILINNRPGPRE PPPYQEPRPRGNPPHSAPCVPNGSALLLSNPAYRLLLATYARPPRGPGPPTPAWAKPTNT QAYSGDYMEPEKPGAPLLPPPPQNSVPHYAEADIVTLQGVTGGNTYAVPALPPGAVGDGP PRVDFPRSRLRFKEKLGEGQFGEVHLCEVDSPQDLVSLDFPLNVRKGHPLLVAVKILRPD ATKNARNDFLKEVKIMSRLKDPNIIRLLGVCVQDDPLCMITDYMENGDLNQFLSAHQLED KAAEGAPGDGQAAQGPTISYPMLLHVAAQIASGMRYLATLNFVHRDLATRNCLVGENFTI KIADFGMSRNLYAGDYYRVQGRAVLPIRWMAWECILMGKFTTASDVWAFGVTLWEVLMLC RAQPFGQLTDEQVIENAGEFFRDQGRQVYLSRPPACPQGLYELMLRCWSRESEQRPPFSQ LHRFLAEDALNTV |
|||||
| Target Type |
Patented-recorded
|
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Protein kinase superfamily, Tyr protein kinase family, Insulin receptor subfamily
|
|||||
| Subcellular location |
Secreted; Cell membrane
|
|||||
| Function |
Tyrosine kinase that functions as a cell surface receptor for fibrillar collagen and regulates cell attachment to the extracellular matrix, remodeling of the extracellular matrix, cell migration, differentiation, survival and cell proliferation. Collagen binding triggers a signaling pathway that involves SRC and leads to the activation of MAP kinases. Regulates remodeling of the extracellular matrix by up-regulation of the matrix metalloproteinases MMP2, MMP7 and MMP9, and thereby facilitates cell migration and wound healing. Required for normal blastocyst implantation during pregnancy, for normal mammary gland differentiation and normal lactation. Required for normal ear morphology and normal hearing. Promotes smooth muscle cell migration, and thereby contributes to arterial wound healing. Also plays a role in tumor cell invasion. Phosphorylates PTPN11.
|
|||||
| TTD ID | ||||||
| Uniprot ID | ||||||
| DrugMap ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
| ChEMBL ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| COLO679 | SNV: p.F241L | . | |||
| HCT15 | SNV: p.I433V | . | |||
| HEC1 | SNV: p.T292M | DBIA Probe Info | |||
| HT115 | SNV: p.M130K; p.K133E | . | |||
| Ishikawa (Heraklio) 02 ER | SNV: p.G131D | . | |||
| KARPAS299 | SNV: p.D795E | . | |||
| KMRC20 | SNV: p.P43T | . | |||
| KMS11 | SNV: p.P806L | . | |||
| NCIH1155 | SNV: p.Q749Ter | . | |||
| NCIH82 | SNV: p.R444C | . | |||
| NUGC3 | Deletion: p.T531HfsTer122 | DBIA Probe Info | |||
| OCIAML5 | SNV: p.S360R | . | |||
| OVK18 | Deletion: p.P483RfsTer170 | . | |||
| RKO | SNV: p.A59G | . | |||
| SHP77 | SNV: p.M311V | . | |||
| SW1783 | SNV: p.R525Q | . | |||
| TE4 | SNV: p.R440L | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STPyne Probe Info |
 |
K646(1.23) | LDD0277 | [1] | |
|
OPA-S-S-alkyne Probe Info |
 |
K30(1.33); K112(2.03) | LDD3494 | [2] | |
|
DBIA Probe Info |
 |
C778(1.59) | LDD3310 | [3] | |
|
JZ128-DTB Probe Info |
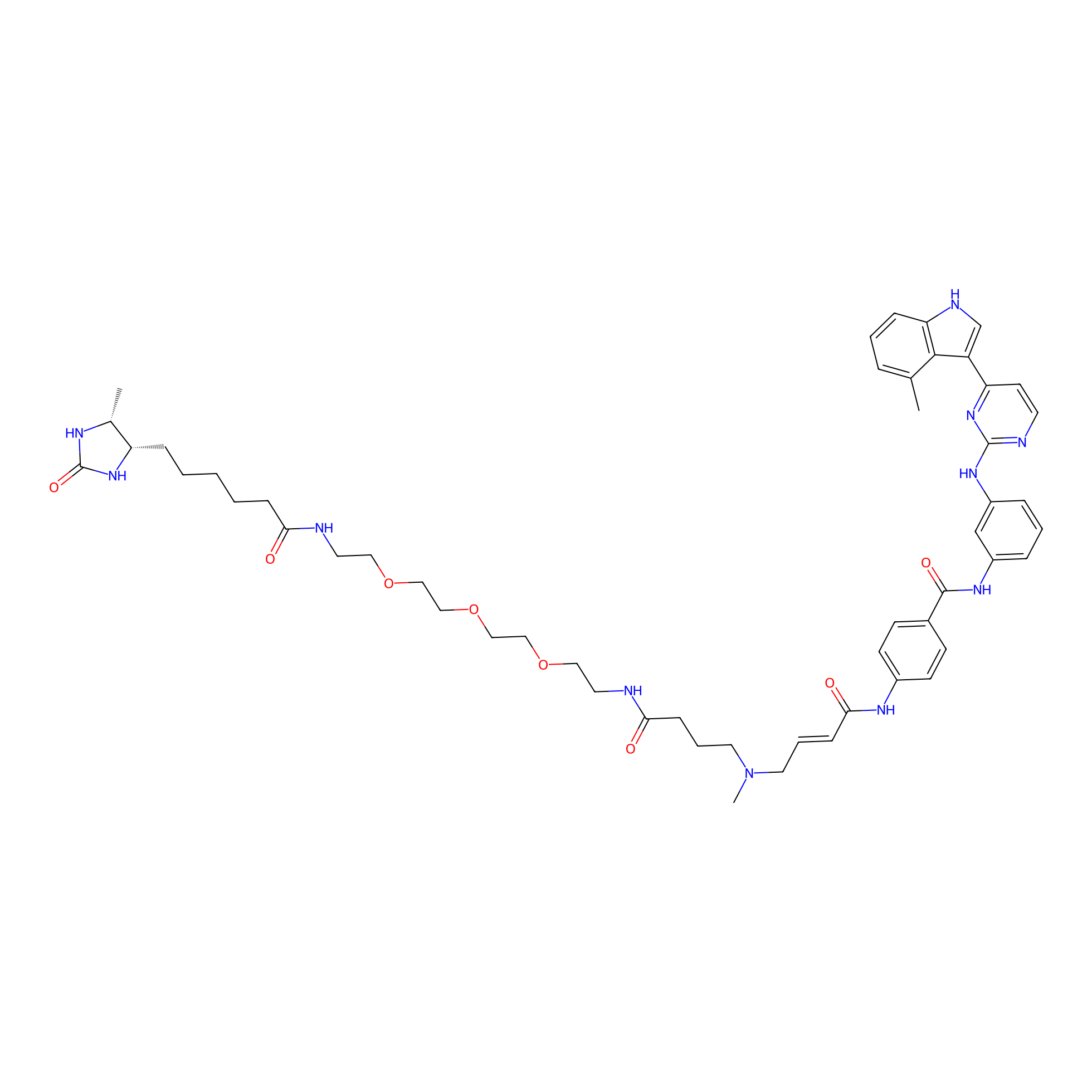 |
N.A. | LDD0462 | [4] | |
|
IA-alkyne Probe Info |
 |
C627(0.00); C772(0.00) | LDD0162 | [5] | |
|
IPM Probe Info |
 |
N.A. | LDD0147 | [6] | |
|
TFBX Probe Info |
 |
N.A. | LDD0148 | [6] | |
|
NAIA_5 Probe Info |
 |
N.A. | LDD2223 | [7] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
Dasatinib-CA-8PAP Probe Info |
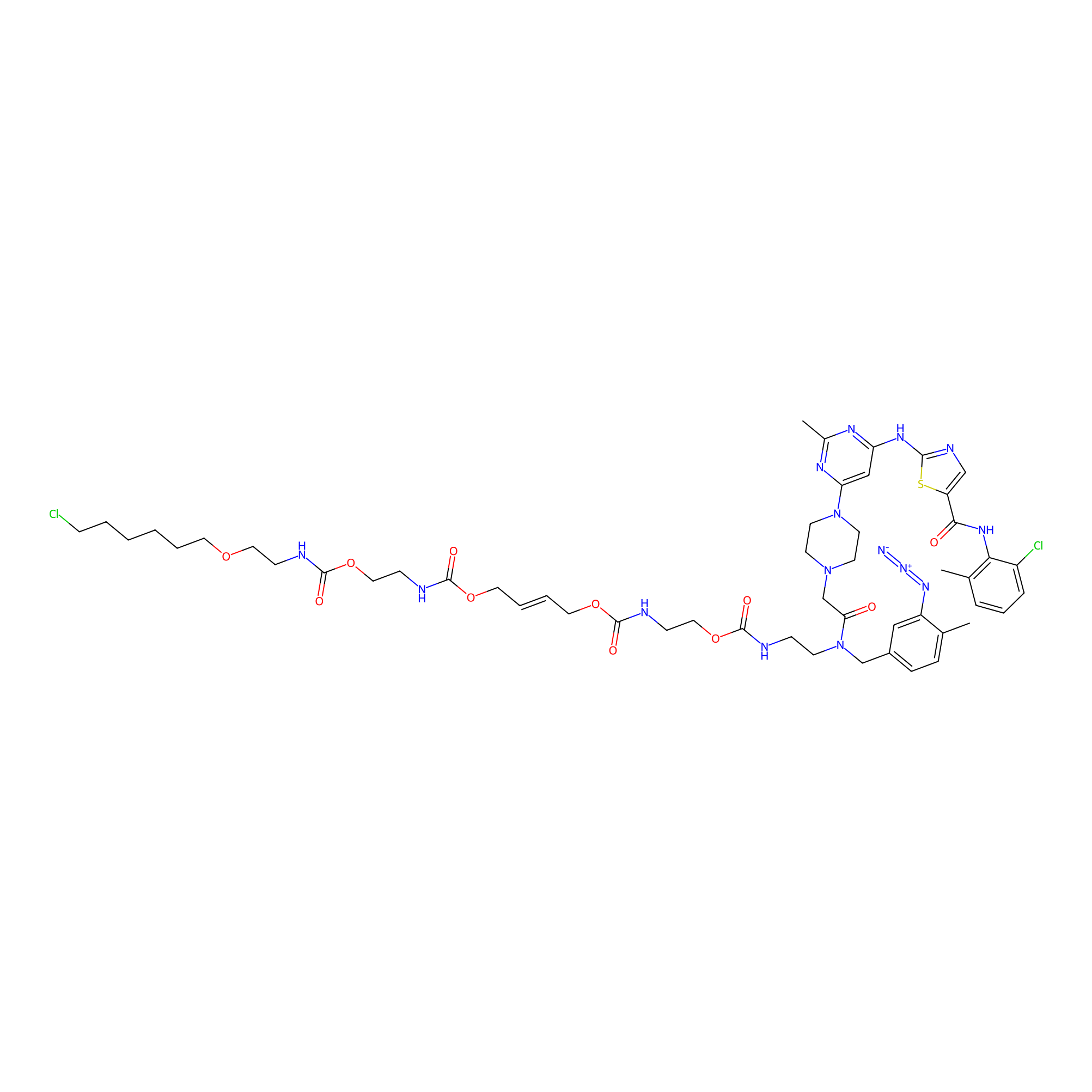 |
N.A. | LDD0364 | [8] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0214 | AC1 | HEK-293T | C876(1.04) | LDD1507 | [9] |
| LDCM0237 | AC12 | HEK-293T | C772(1.31) | LDD1510 | [9] |
| LDCM0276 | AC17 | HEK-293T | C876(1.11) | LDD1515 | [9] |
| LDCM0280 | AC20 | HEK-293T | C772(0.95) | LDD1519 | [9] |
| LDCM0285 | AC25 | HEK-293T | C876(0.90) | LDD1524 | [9] |
| LDCM0288 | AC28 | HEK-293T | C772(1.11) | LDD1527 | [9] |
| LDCM0294 | AC33 | HEK-293T | C876(0.96) | LDD1533 | [9] |
| LDCM0297 | AC36 | HEK-293T | C772(1.17) | LDD1536 | [9] |
| LDCM0301 | AC4 | HEK-293T | C772(1.53) | LDD1540 | [9] |
| LDCM0303 | AC41 | HEK-293T | C876(1.16) | LDD1542 | [9] |
| LDCM0306 | AC44 | HEK-293T | C772(1.03) | LDD1545 | [9] |
| LDCM0311 | AC49 | HEK-293T | C876(0.97) | LDD1550 | [9] |
| LDCM0315 | AC52 | HEK-293T | C772(1.33) | LDD1554 | [9] |
| LDCM0320 | AC57 | HEK-293T | C876(1.09) | LDD1559 | [9] |
| LDCM0324 | AC60 | HEK-293T | C772(1.07) | LDD1563 | [9] |
| LDCM0356 | AKOS034007680 | HEK-293T | C876(0.88) | LDD1570 | [9] |
| LDCM0404 | CL17 | HEK-293T | C876(1.24) | LDD1608 | [9] |
| LDCM0408 | CL20 | HEK-293T | C772(1.42) | LDD1612 | [9] |
| LDCM0417 | CL29 | HEK-293T | C876(1.00) | LDD1621 | [9] |
| LDCM0421 | CL32 | HEK-293T | C772(1.24) | LDD1625 | [9] |
| LDCM0431 | CL41 | HEK-293T | C876(1.17) | LDD1635 | [9] |
| LDCM0434 | CL44 | HEK-293T | C772(1.27) | LDD1638 | [9] |
| LDCM0440 | CL5 | HEK-293T | C876(1.02) | LDD1644 | [9] |
| LDCM0444 | CL53 | HEK-293T | C876(1.18) | LDD1647 | [9] |
| LDCM0447 | CL56 | HEK-293T | C772(1.67) | LDD1650 | [9] |
| LDCM0457 | CL65 | HEK-293T | C876(1.27) | LDD1660 | [9] |
| LDCM0460 | CL68 | HEK-293T | C772(1.12) | LDD1663 | [9] |
| LDCM0470 | CL77 | HEK-293T | C876(1.67) | LDD1673 | [9] |
| LDCM0473 | CL8 | HEK-293T | C772(1.29) | LDD1676 | [9] |
| LDCM0474 | CL80 | HEK-293T | C772(1.13) | LDD1677 | [9] |
| LDCM0483 | CL89 | HEK-293T | C876(0.99) | LDD1686 | [9] |
| LDCM0487 | CL92 | HEK-293T | C772(1.15) | LDD1690 | [9] |
| LDCM0097 | Dasatinib | K562 | N.A. | LDD0364 | [8] |
| LDCM0179 | JZ128 | PC-3 | N.A. | LDD0462 | [4] |
| LDCM0022 | KB02 | 22RV1 | C305(2.14); C627(0.73); C778(2.44); C882(2.43) | LDD2243 | [3] |
| LDCM0023 | KB03 | 22RV1 | C305(2.55); C627(1.03); C778(4.01); C882(2.74) | LDD2660 | [3] |
| LDCM0024 | KB05 | COLO792 | C778(1.59) | LDD3310 | [3] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| Cytoplasmic protein NCK2 (NCK2) | . | O43639 | |||
| Ubiquilin-2 (UBQLN2) | . | Q9UHD9 | |||
The Drug(s) Related To This Target
Approved
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Fostamatinib | Small molecular drug | DB12010 | |||
| Imatinib | Small molecular drug | DB00619 | |||
| Pralsetinib | . | DB15822 | |||
Investigative
| Drug Name | Drug Type | External ID | |||
|---|---|---|---|---|---|
| Pmid23521020c7k | Small molecular drug | D0BL0O | |||
Patented
References
