Details of the Target
General Information of Target
| Target ID | LDTP15391 | |||||
|---|---|---|---|---|---|---|
| Target Name | Probable phosphoglycerate mutase 4 (PGAM4) | |||||
| Gene Name | PGAM4 | |||||
| Gene ID | 441531 | |||||
| Synonyms |
PGAM3; Probable phosphoglycerate mutase 4; EC 5.4.2.11; EC 5.4.2.4 |
|||||
| 3D Structure | ||||||
| Sequence |
MALPQMCDGSHLASTLRYCMTVSGTVVLVAGTLCFAWWSEGDATAQPGQLAPPTEYPVPE
GPSPLLRSVSFVCCGAGGLLLLIGLLWSVKASIPGPPRWDPYHLSRDLYYLTVESSEKES CRTPKVVDIPTYEEAVSFPVAEGPPTPPAYPTEEALEPSGSRDALLSTQPAWPPPSYESI SLALDAVSAETTPSATRSCSGLVQTARGGS |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
Phosphoglycerate mutase family, BPG-dependent PGAM subfamily
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
THZ1-DTB Probe Info |
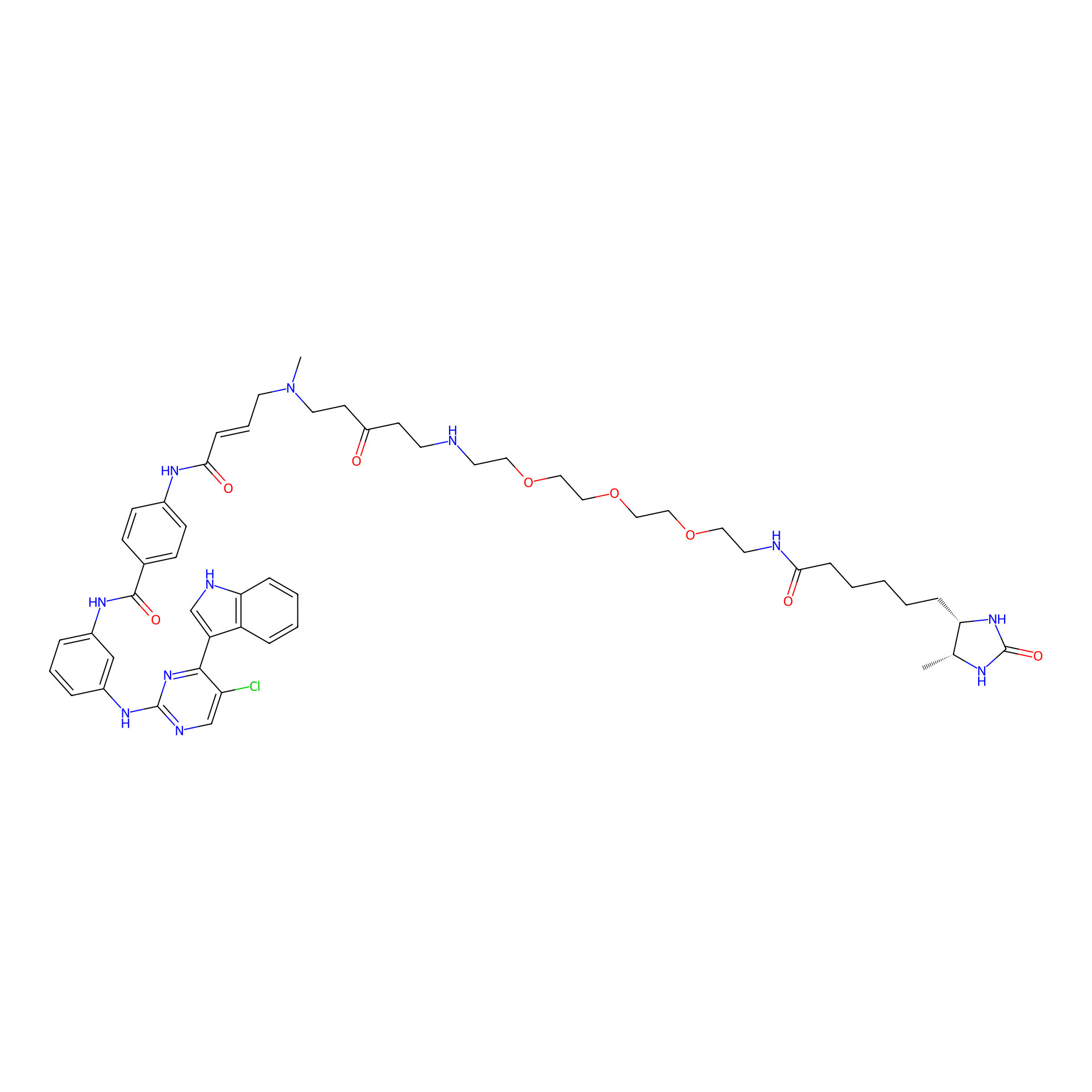 |
C55(1.00) | LDD0460 | [1] | |
|
HHS-475 Probe Info |
 |
Y133(0.74); Y119(0.84); Y92(0.97) | LDD0264 | [2] | |
|
HHS-465 Probe Info |
 |
Y119(4.96); Y133(7.45); Y92(10.00) | LDD2237 | [3] | |
|
Acrolein Probe Info |
 |
N.A. | LDD0217 | [4] | |
|
AOyne Probe Info |
 |
6.60 | LDD0443 | [5] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DA-2 Probe Info |
 |
N.A. | LDD0072 | [6] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0116 | HHS-0101 | DM93 | Y133(0.74); Y119(0.84); Y92(0.97) | LDD0264 | [2] |
| LDCM0117 | HHS-0201 | DM93 | Y133(0.70); Y119(0.81); Y92(0.90) | LDD0265 | [2] |
| LDCM0118 | HHS-0301 | DM93 | Y133(0.81); Y92(0.89); Y119(0.99) | LDD0266 | [2] |
| LDCM0119 | HHS-0401 | DM93 | Y133(0.80); Y92(1.06); Y119(5.68) | LDD0267 | [2] |
| LDCM0120 | HHS-0701 | DM93 | Y133(0.88); Y119(1.14); Y92(1.18) | LDD0268 | [2] |
| LDCM0107 | IAA | HeLa | N.A. | LDD0221 | [4] |
| LDCM0109 | NEM | HeLa | N.A. | LDD0223 | [4] |
| LDCM0021 | THZ1 | HeLa S3 | C55(1.00) | LDD0460 | [1] |
References
