Details of the Target
General Information of Target
| Target ID | LDTP13618 | |||||
|---|---|---|---|---|---|---|
| Target Name | ATP-dependent RNA helicase DDX19B (DDX19B) | |||||
| Gene Name | DDX19B | |||||
| Gene ID | 11269 | |||||
| Synonyms |
DBP5; DDX19; TDBP; ATP-dependent RNA helicase DDX19B; EC 3.6.4.13; DEAD box RNA helicase DEAD5; DEAD box protein 19B |
|||||
| 3D Structure | ||||||
| Sequence |
MLYSPGPSLPESAESLDGSQEDKPRGSCAEPTFTDTGMVAHINNSRLKAKGVGQHDNAQN
FGNQSFEELRAACLRKGELFEDPLFPAEPSSLGFKDLGPNSKNVQNISWQRPKDIINNPL FIMDGISPTDICQGILGDCWLLAAIGSLTTCPKLLYRVVPRGQSFKKNYAGIFHFQIWQF GQWVNVVVDDRLPTKNDKLVFVHSTERSEFWSALLEKAYAKLSGSYEALSGGSTMEGLED FTGGVAQSFQLQRPPQNLLRLLRKAVERSSLMGCSIEVTSDSELESMTDKMLVRGHAYSV TGLQDVHYRGKMETLIRVRNPWGRIEWNGAWSDSAREWEEVASDIQMQLLHKTEDGEFWM SYQDFLNNFTLLEICNLTPDTLSGDYKSYWHTTFYEGSWRRGSSAGGCRNHPGTFWTNPQ FKISLPEGDDPEDDAEGNVVVCTCLVALMQKNWRHARQQGAQLQTIGFVLYAVPKEFQNI QDVHLKKEFFTKYQDHGFSEIFTNSREVSSQLRLPPGEYIIIPSTFEPHRDADFLLRVFT EKHSESWELDEVNYAEQLQEEKVSEDDMDQDFLHLFKIVAGEGKEIGVYELQRLLNRMAI KFKSFKTKGFGLDACRCMINLMDKDGSGKLGLLEFKILWKKLKKWMDIFRECDQDHSGTL NSYEMRLVIEKAGIKLNNKVMQVLVARYADDDLIIDFDSFISCFLRLKTMFTFFLTMDPK NTGHICLSLEQWLQMTMWG |
|||||
| Target Bioclass |
Enzyme
|
|||||
| Family |
DEAD box helicase family, DDX19/DBP5 subfamily
|
|||||
| Subcellular location |
Cytoplasm
|
|||||
| Function |
ATP-dependent RNA helicase involved in mRNA export from the nucleus. Rather than unwinding RNA duplexes, DDX19B functions as a remodeler of ribonucleoprotein particles, whereby proteins bound to nuclear mRNA are dissociated and replaced by cytoplasmic mRNA binding proteins.
|
|||||
| Uniprot ID | ||||||
| Ensemble ID | ||||||
| HGNC ID | ||||||
Target Site Mutations in Different Cell Lines
| Cell line | Mutation details | Probe for labeling this protein in this cell | |||
|---|---|---|---|---|---|
| DOTC24510 | SNV: p.Q322Ter | DBIA Probe Info | |||
| LNCaP clone FGC | SNV: p.P84S | DBIA Probe Info | |||
| LS123 | SNV: p.A346G | DBIA Probe Info | |||
| LS180 | Deletion: p.K385del | DBIA Probe Info | |||
| NCIH1155 | SNV: p.Q13Ter; p.V281M | DBIA Probe Info | |||
| NCIH2170 | SNV: p.G106R | DBIA Probe Info | |||
| RL952 | Substitution: p.H24_L25delinsQF | DBIA Probe Info | |||
| SNU1196 | SNV: p.E183Q | DBIA Probe Info | |||
Probe(s) Labeling This Target
ABPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
DBIA Probe Info |
 |
C314(3.48); C393(3.04) | LDD3312 | [1] | |
|
THZ1-DTB Probe Info |
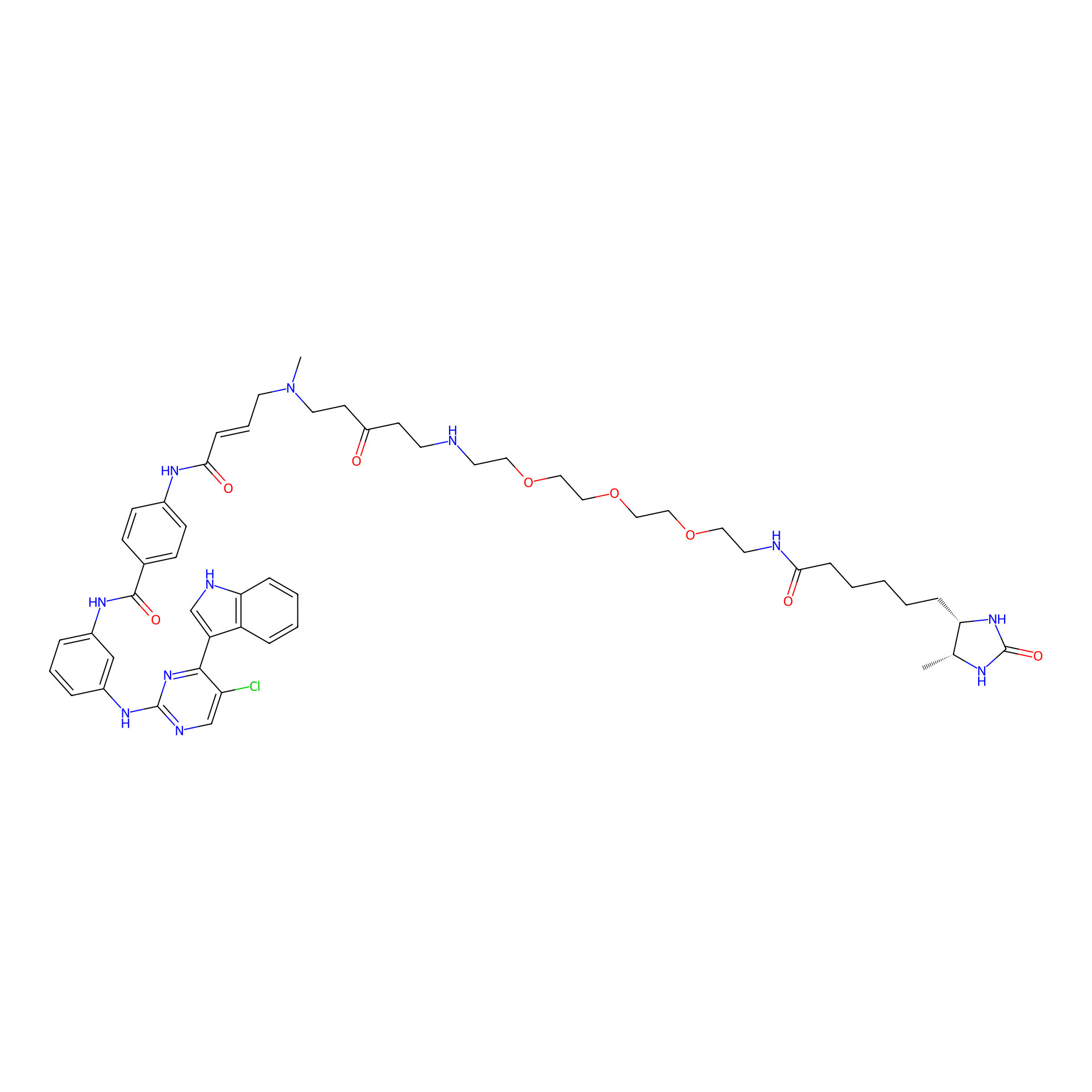 |
C225(1.07) | LDD0460 | [2] | |
|
BTD Probe Info |
 |
C393(0.41) | LDD2089 | [3] | |
|
AHL-Pu-1 Probe Info |
 |
C393(2.67) | LDD0168 | [4] | |
|
HHS-475 Probe Info |
 |
Y89(0.83) | LDD0264 | [5] | |
|
IA-alkyne Probe Info |
 |
C393(0.00); C225(0.00) | LDD0032 | [6] | |
|
ATP probe Probe Info |
 |
N.A. | LDD0035 | [7] | |
|
NAIA_4 Probe Info |
 |
N.A. | LDD2226 | [8] | |
|
TFBX Probe Info |
 |
N.A. | LDD0027 | [9] | |
|
WYneN Probe Info |
 |
C225(0.00); C393(0.00) | LDD0021 | [10] | |
|
WYneO Probe Info |
 |
N.A. | LDD0022 | [10] | |
|
IPM Probe Info |
 |
N.A. | LDD0005 | [10] | |
|
NHS Probe Info |
 |
N.A. | LDD0010 | [10] | |
|
STPyne Probe Info |
 |
N.A. | LDD0009 | [10] | |
|
Phosphinate-6 Probe Info |
 |
N.A. | LDD0018 | [11] | |
|
Ox-W18 Probe Info |
 |
N.A. | LDD2175 | [12] | |
|
AOyne Probe Info |
 |
10.30 | LDD0443 | [13] | |
|
HHS-465 Probe Info |
 |
K92(0.00); Y89(0.00) | LDD2240 | [14] | |
PAL-AfBPP Probe
| Probe name | Structure | Binding Site(Ratio) | Interaction ID | Ref | |
|---|---|---|---|---|---|
|
STS-2 Probe Info |
 |
N.A. | LDD0138 | [15] | |
Competitor(s) Related to This Target
| Competitor ID | Name | Cell line | Binding Site(Ratio) | Interaction ID | Ref |
|---|---|---|---|---|---|
| LDCM0524 | 2-Cyano-N-(2-morpholin-4-yl-ethyl)-acetamide | MDA-MB-231 | C393(0.91) | LDD2117 | [3] |
| LDCM0558 | 2-Cyano-N-phenylacetamide | MDA-MB-231 | C393(0.91) | LDD2152 | [3] |
| LDCM0025 | 4SU-RNA | HEK-293T | C393(2.67) | LDD0168 | [4] |
| LDCM0026 | 4SU-RNA+native RNA | HEK-293T | C393(2.00) | LDD0169 | [4] |
| LDCM0545 | Acetamide | MDA-MB-231 | C393(0.51) | LDD2138 | [3] |
| LDCM0520 | AKOS000195272 | MDA-MB-231 | C393(0.99) | LDD2113 | [3] |
| LDCM0020 | ARS-1620 | HCC44 | C393(0.96) | LDD2171 | [16] |
| LDCM0498 | BS-3668 | MDA-MB-231 | C393(0.40) | LDD2091 | [3] |
| LDCM0630 | CCW28-3 | 231MFP | C393(1.44) | LDD2214 | [17] |
| LDCM0213 | Electrophilic fragment 2 | MDA-MB-231 | C225(2.52) | LDD1702 | [3] |
| LDCM0116 | HHS-0101 | DM93 | Y89(0.83) | LDD0264 | [5] |
| LDCM0117 | HHS-0201 | DM93 | Y89(0.58) | LDD0265 | [5] |
| LDCM0118 | HHS-0301 | DM93 | Y89(0.34) | LDD0266 | [5] |
| LDCM0119 | HHS-0401 | DM93 | Y89(1.01) | LDD0267 | [5] |
| LDCM0120 | HHS-0701 | DM93 | Y89(1.53) | LDD0268 | [5] |
| LDCM0022 | KB02 | 22RV1 | C314(1.52); C393(2.03) | LDD2243 | [1] |
| LDCM0023 | KB03 | MDA-MB-231 | C393(6.86) | LDD1701 | [3] |
| LDCM0024 | KB05 | HMCB | C314(3.48); C393(3.04) | LDD3312 | [1] |
| LDCM0528 | N-(4-bromophenyl)-2-cyano-N-phenylacetamide | MDA-MB-231 | C393(0.50) | LDD2121 | [3] |
| LDCM0496 | Nucleophilic fragment 11a | MDA-MB-231 | C393(0.41) | LDD2089 | [3] |
| LDCM0497 | Nucleophilic fragment 11b | MDA-MB-231 | C393(1.17) | LDD2090 | [3] |
| LDCM0500 | Nucleophilic fragment 13a | MDA-MB-231 | C393(0.89) | LDD2093 | [3] |
| LDCM0514 | Nucleophilic fragment 20a | MDA-MB-231 | C393(0.94) | LDD2107 | [3] |
| LDCM0516 | Nucleophilic fragment 21a | MDA-MB-231 | C393(0.52) | LDD2109 | [3] |
| LDCM0518 | Nucleophilic fragment 22a | MDA-MB-231 | C393(0.86) | LDD2111 | [3] |
| LDCM0522 | Nucleophilic fragment 24a | MDA-MB-231 | C393(0.92) | LDD2115 | [3] |
| LDCM0525 | Nucleophilic fragment 25b | MDA-MB-231 | C393(0.67) | LDD2118 | [3] |
| LDCM0527 | Nucleophilic fragment 26b | MDA-MB-231 | C393(1.08) | LDD2120 | [3] |
| LDCM0530 | Nucleophilic fragment 28a | MDA-MB-231 | C393(0.73) | LDD2123 | [3] |
| LDCM0531 | Nucleophilic fragment 28b | MDA-MB-231 | C393(0.66) | LDD2124 | [3] |
| LDCM0532 | Nucleophilic fragment 29a | MDA-MB-231 | C393(0.59) | LDD2125 | [3] |
| LDCM0536 | Nucleophilic fragment 31 | MDA-MB-231 | C393(0.91) | LDD2129 | [3] |
| LDCM0541 | Nucleophilic fragment 36 | MDA-MB-231 | C393(0.65) | LDD2134 | [3] |
| LDCM0542 | Nucleophilic fragment 37 | MDA-MB-231 | C393(1.04) | LDD2135 | [3] |
| LDCM0543 | Nucleophilic fragment 38 | MDA-MB-231 | C393(1.02) | LDD2136 | [3] |
| LDCM0546 | Nucleophilic fragment 40 | MDA-MB-231 | C393(0.55) | LDD2140 | [3] |
| LDCM0552 | Nucleophilic fragment 6a | MDA-MB-231 | C393(0.80) | LDD2146 | [3] |
| LDCM0554 | Nucleophilic fragment 7a | MDA-MB-231 | C393(0.49) | LDD2148 | [3] |
| LDCM0559 | Nucleophilic fragment 9b | MDA-MB-231 | C393(1.14) | LDD2153 | [3] |
| LDCM0627 | NUDT7-COV-1 | HEK-293T | C393(0.92) | LDD2206 | [18] |
| LDCM0628 | OTUB2-COV-1 | HEK-293T | C393(0.76) | LDD2207 | [18] |
| LDCM0021 | THZ1 | HeLa S3 | C225(1.07) | LDD0460 | [2] |
The Interaction Atlas With This Target
The Protein(s) Related To This Target
Enzyme
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| TGF-beta receptor type-2 (TGFBR2) | TKL Ser/Thr protein kinase family | P37173 | |||
Other
| Protein name | Family | Uniprot ID | |||
|---|---|---|---|---|---|
| MIF4G domain-containing protein (MIF4GD) | MIF4GD family | A9UHW6 | |||
| Chromobox protein homolog 5 (CBX5) | . | P45973 | |||
| Telomeric repeat-binding factor 1 (TERF1) | . | P54274 | |||
References
